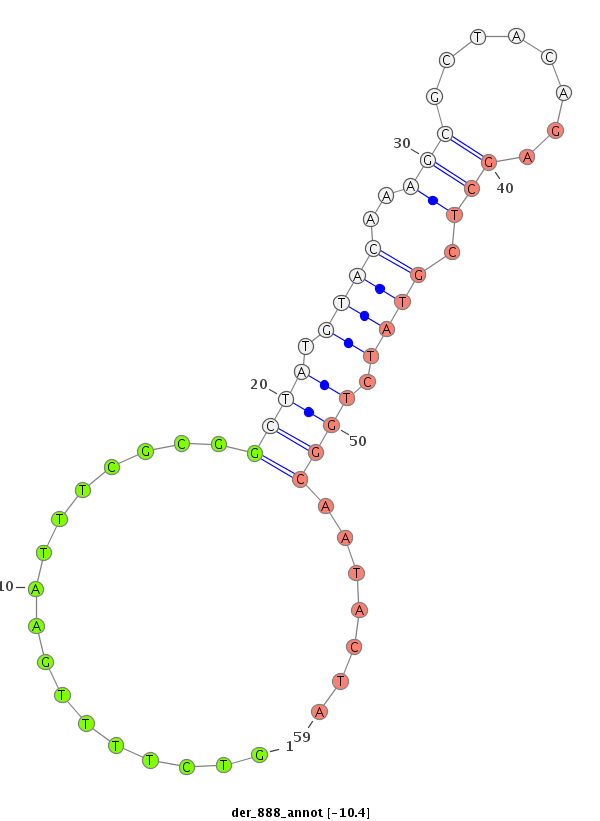
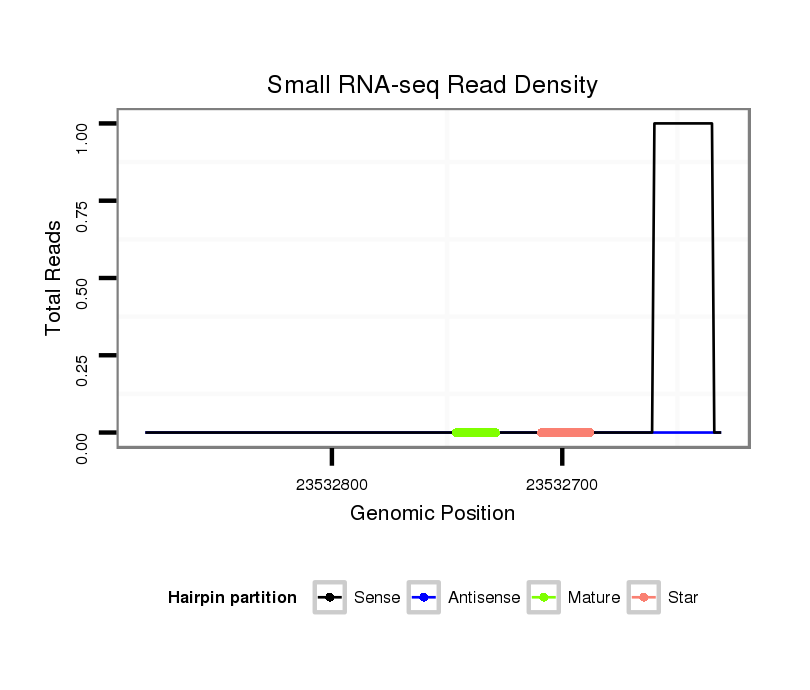
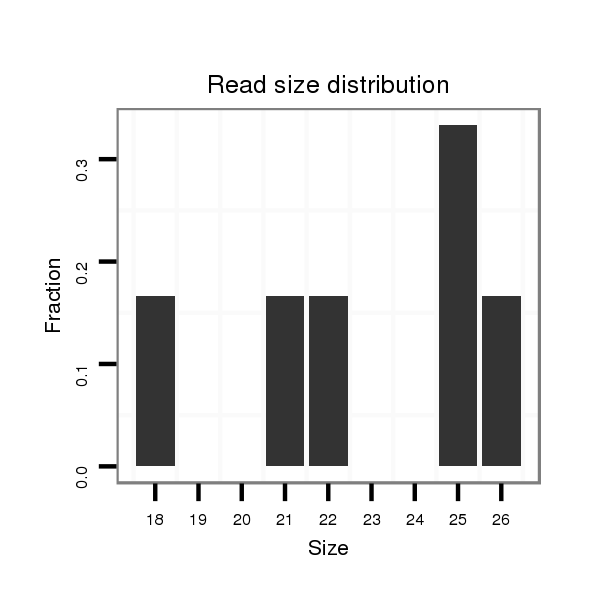
| TGCAAAAAA----TTGGT--------------------------------------------------------------AAATTGGT----------A------------------------------------------------------------------------------TTGGACTATATATTGAGT-------------------------TA----------------------------------TTTTTC----------------CAG-ATATAAG-------------------C----------GAT-----------------------ATATAAGGTTG------TCAAATA-------------------------------------T---------CT-----CCTTTC-----CGCCT--TTT---------TGTC-TTTTGAATTTCGCGGCT-----------ATG--TACAA-AGCGCTACAGAGCTTGTATCTGGCAATACTATACATAATTTGAAA-GGTCTTGGCATAACCTACAAAACGACGCTATG----------------------ATTAGTTTG-------------GA | Size | Hit Count | Total Norm | Total | M044
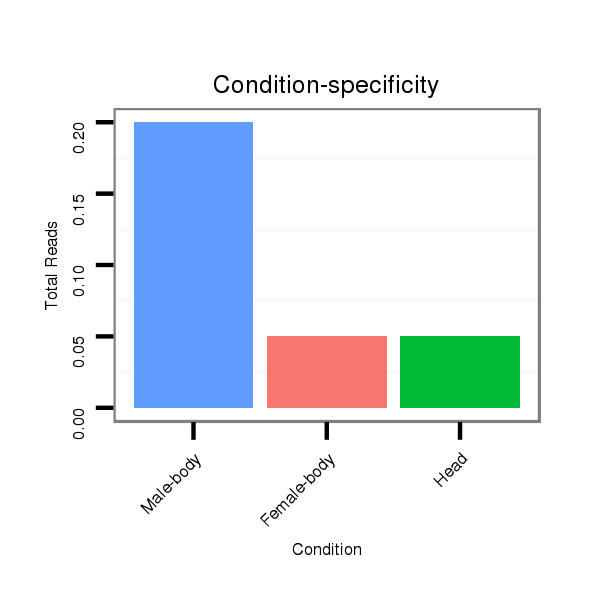
Female-body | M058
Embryo | V039
Embryo | V055
Head | V105
Male-body | V106
Head |
| .......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TTTGAAA-GGTCTTGGCATAACCTACAA.......................................................... | 27 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TTTGAAA-GGTCTTGGCATAACCTACA........................................................... | 26 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TACATAATTTGAAA-GGTCTTGGC..................................................................... | 23 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TACATAATTTGAAA-GGTCTTGGCATAAC................................................................ | 28 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TACAAAACGACGCTATG----------------------ATTAGTT................. | 24 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TACAAAACGACGCTATG----------------------ATTAGTTTG-------------.. | 26 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................CCTACAAAACGACGCTATG----------------------ATTAGTTTG-------------.. | 28 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................ACATAATTTGAAA-GGTCTTGGCA.................................................................... | 23 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................................................................................................................................C-TTTTGAATTTCGCGGCT-----------ATG--TACA.............................................................................................................................. | 25 | 20 | 0.45 | 9 | 9 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................................................................................................................................TTTTGAATTTCGCGGCT-----------ATG--TACA.............................................................................................................................. | 24 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................................................................................................................................................................................TGAATTTCGCGGCT-----------ATG--TACAA-AGCGC....................................................................................................................... | 27 | 17 | 0.12 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................................................................................................................................TC-TTTTGAATTTCGCGGCT-----------ATG--TACA.............................................................................................................................. | 26 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................................................................................................................................TC-TTTTGAATTTCGCGGCT-----------ATG--TAC............................................................................................................................... | 25 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................................................................................................................................TC-TTTTGAATTTCGCGGCT-----------ATG--TA................................................................................................................................ | 24 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TTCGCGGCT-----------ATG--TACAA-AGCGCTACAG.................................................................................................................. | 27 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................................................................................................................................TTTTGAATTTCGCGGCT-----------ATG--TACAA-............................................................................................................................ | 25 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................................................................................................................................................TTGAATTTCGCGGCT-----------ATG--TACAA-A........................................................................................................................... | 24 | 17 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................G--TACAA-AGCGCTACAGAGCT.............................................................................................................. | 20 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................................................................................................................................TTTTGAATTTCGCGGCT-----------ATG--TACAA-AGC......................................................................................................................... | 28 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................................................................................................................................................................TGTC-TTTTGAATTTCGCGGCT-----------ATG--TAC............................................................................................................................... | 27 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TACTATACATAATTTGAAA-GGTCTTG....................................................................... | 26 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................................................................................................................................................GTC-TTTTGAATTTCGCGGCT-----------ATG--TAC............................................................................................................................... | 26 | 18 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................................TACATAATTTGAAA-GGTCTTG....................................................................... | 21 | 19 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................................................................................................................................................................TGTC-TTTTGAATTTCGCGGCT-----------ATG--TA................................................................................................................................ | 26 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................................................................................................................................C-TTTTGAATTTCGCGGCT-----------ATG--TAC............................................................................................................................... | 24 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................................................................................................................................................................TC-TTTTGAATTTCGCGGCT-----------ATG--T................................................................................................................................. | 23 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |