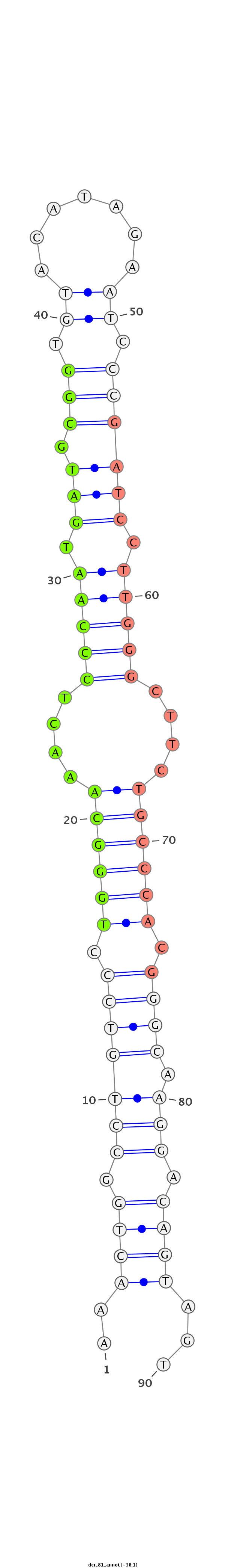
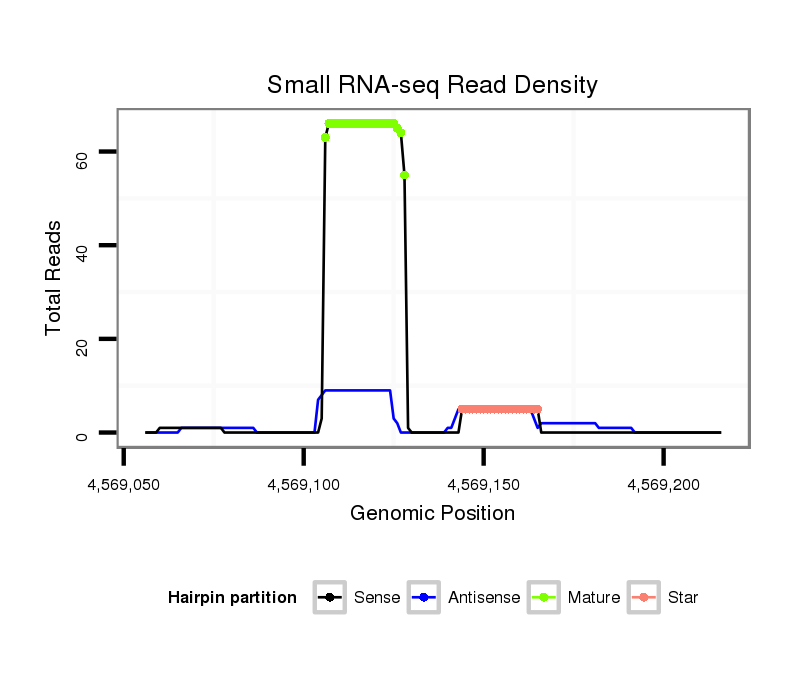
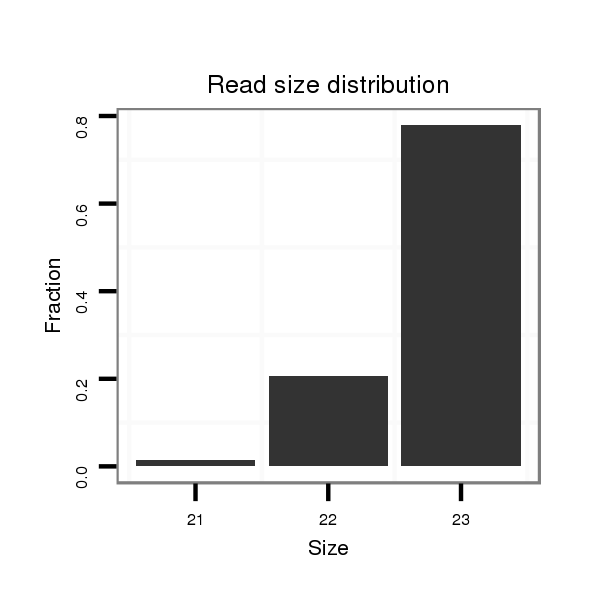
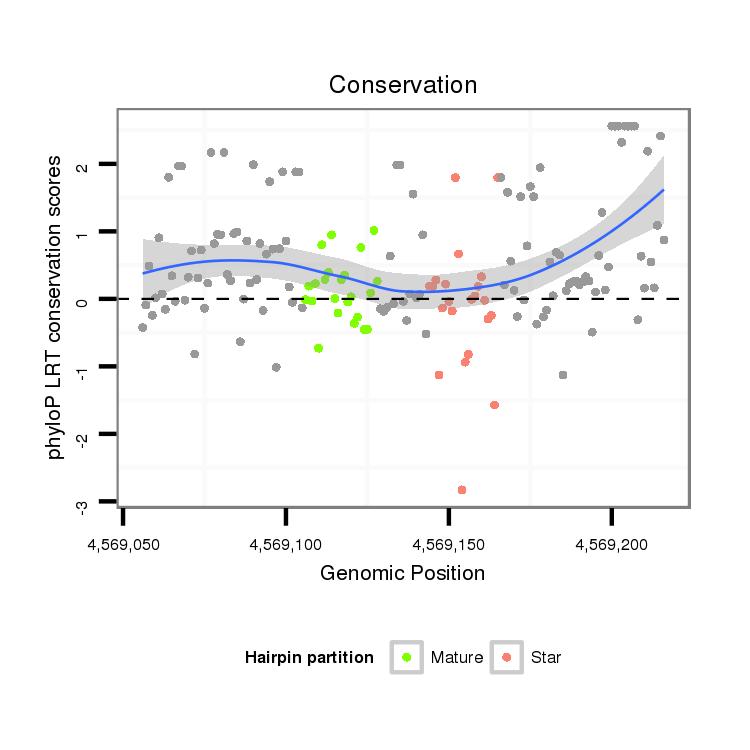
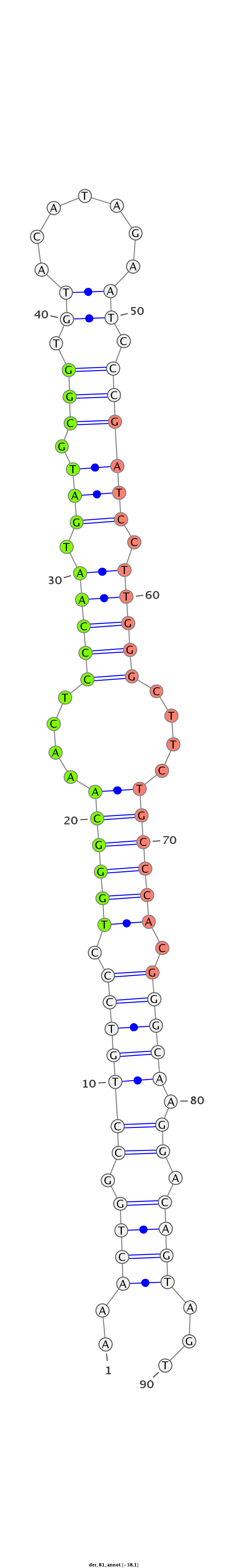
| CGAGG---------------------GT-----------------------------TAG-CCCGC------------GATGATGTAAGCTGGCCTGTCCCTGGGTAA--ACTCCC----------------------------------------------------------------------------AAGGAT-----------------------CAGGTGGAGATTGAA-----------------T------------------CCCGATCCTTGGGATTCTGCTCTCGGGCA-AGGTCAGCAGTGCCCG--CGAAG--------GGCCCAGCTGTTTGTTAACTCC--------TTTAG | Size | Hit Count | Total Norm | Total | M054
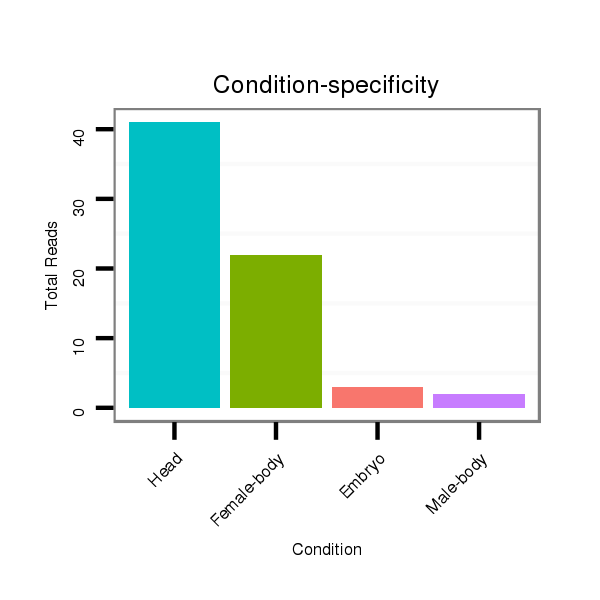
Female-body | V113
Male-body | V114
Embryo | V115
Head |
| .....................................................................................................TGGGTAA--ACTCCC----------------------------------------------------------------------------AAGGAT-----------------------CAGG............................................................................................................................................... | 23 | 1 | 913.00 | 913 | 128 | 173 | 67 | 545 |
| .....................................................................................................TGGGTAA--ACTCCC----------------------------------------------------------------------------AAGGAT-----------------------CAG................................................................................................................................................ | 22 | 1 | 595.00 | 595 | 90 | 135 | 37 | 333 |
| .....................................................................................................TGGGTAA--ACTCCC----------------------------------------------------------------------------AAGGAT-----------------------CA................................................................................................................................................. | 21 | 1 | 192.00 | 192 | 51 | 12 | 7 | 122 |
| ..................................................................................................................................................................................................................................................................................CGATCCTTGGGATTCTGCTCTCGG...................................................................... | 24 | 1 | 168.00 | 168 | 74 | 17 | 22 | 55 |
| ....................................................................................................................................................................................................................................................................................ATCCTTGGGATTCTGCTCTCGG...................................................................... | 22 | 1 | 141.00 | 141 | 99 | 11 | 2 | 29 |
| .....................................................................................................TGGGTAA--ACTCCC----------------------------------------------------------------------------AAGGAT-----------------------CAGGT.............................................................................................................................................. | 24 | 1 | 90.00 | 90 | 4 | 42 | 0 | 44 |
| ....................................................................................................................................................................................................................................................................................ATCCTTGGGATTCTGCTCTCG....................................................................... | 21 | 1 | 85.00 | 85 | 15 | 32 | 1 | 37 |
| ..................................................................................................................................................................................................................................................................................CGATCCTTGGGATTCTGCTCTCG....................................................................... | 23 | 1 | 82.00 | 82 | 22 | 15 | 5 | 40 |
| .....................................................................................................TGGGTAA--ACTCCC----------------------------------------------------------------------------AAGGAT-----------------------C.................................................................................................................................................. | 20 | 1 | 63.00 | 63 | 24 | 0 | 1 | 38 |
| ....................................................................................................................................................................................................................................................................................ATCCTTGGGATTCTGCTCTC........................................................................ | 20 | 1 | 51.00 | 51 | 12 | 5 | 0 | 34 |
| .....................................................................................................TGGGTAA--ACTCCC----------------------------------------------------------------------------AAGGAT-----------------------CAGT............................................................................................................................................... | 23 | 1 | 27.00 | 27 | 4 | 15 | 0 | 8 |
| .....................................................................................................TGGGTAA--ACTCCC----------------------------------------------------------------------------AAGGAT-----------------------CAGGA.............................................................................................................................................. | 24 | 1 | 20.00 | 20 | 1 | 7 | 1 | 11 |
| ......................................................................................................GGGTAA--ACTCCC----------------------------------------------------------------------------AAGGAT-----------------------CAGG............................................................................................................................................... | 22 | 1 | 16.00 | 16 | 15 | 1 | 0 | 0 |
| .....................................................................................................TGGGTAA--ACTCCC----------------------------------------------------------------------------AAGGAT-----------------------CAGGTT............................................................................................................................................. | 25 | 1 | 16.00 | 16 | 0 | 11 | 0 | 5 |
| ......................................................................................................GGGTAA--ACTCCC----------------------------------------------------------------------------AAGGAT-----------------------CAG................................................................................................................................................ | 21 | 1 | 15.00 | 15 | 15 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................CGATCCTTGGGATTCTGCTCTC........................................................................ | 22 | 1 | 13.00 | 13 | 8 | 1 | 1 | 3 |
| .....................................................................................................TGGGTAA--ACTCCC----------------------------------------------------------------------------AAGGAT-----------------------CAGA............................................................................................................................................... | 23 | 1 | 12.00 | 12 | 2 | 4 | 1 | 5 |
| ...................................................................................................................................................................................................................................................................................GATCCTTGGGATTCTGCTCTCGG...................................................................... | 23 | 1 | 12.00 | 12 | 9 | 0 | 1 | 2 |
| .....................................................................................................................................................................................................................................................................................TCCTTGGGATTCTGCTCTCGG...................................................................... | 21 | 1 | 11.00 | 11 | 8 | 1 | 2 | 0 |
| ...................................................................................................................................................................................................................................................................................GATCCTTGGGATTCTGCTCTCG....................................................................... | 22 | 1 | 9.00 | 9 | 4 | 3 | 0 | 2 |
| ....................................................................................................CTGGGTAA--ACTCCC----------------------------------------------------------------------------AAGGAT-----------------------C.................................................................................................................................................. | 21 | 1 | 7.00 | 7 | 4 | 0 | 0 | 3 |
| ..................................................................................................................................................................................................................................................................................CGATCCTTGGGATTCTGCTCTCGGT..................................................................... | 25 | 1 | 6.00 | 6 | 0 | 1 | 0 | 5 |
| ....................................................................................................................................................................................................................................................................................ATCCTTGGGATTCTGCTCTCGGT..................................................................... | 23 | 1 | 6.00 | 6 | 6 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................CGATCCTTGGGATTCTGCTCT......................................................................... | 21 | 1 | 5.00 | 5 | 4 | 0 | 0 | 1 |
| ...................................................................................................................................................................................................................................................................................GATCCTTGGGATTCTGCTCTC........................................................................ | 21 | 1 | 4.00 | 4 | 4 | 0 | 0 | 0 |
| .....................................................................................................TGGGTAA--ACTCCC----------------------------------------------------------------------------AAGGAT-----------------------CAGGC.............................................................................................................................................. | 24 | 1 | 4.00 | 4 | 0 | 1 | 0 | 3 |
| ....................................................................................................CTGGGTAA--ACTCCC----------------------------------------------------------------------------AAGGAT-----------------------CA................................................................................................................................................. | 22 | 1 | 3.00 | 3 | 1 | 0 | 1 | 1 |
| .......................................................................................................GGTAA--ACTCCC----------------------------------------------------------------------------AAGGAT-----------------------CAG................................................................................................................................................ | 20 | 1 | 3.00 | 3 | 2 | 0 | 0 | 1 |
| ....................................................................................................................................................................................................................................................................................ATCCTTGGGATTCTGCTCTCGT...................................................................... | 22 | 1 | 3.00 | 3 | 1 | 1 | 0 | 1 |
| ..................................................................................................................................................................................................................................................................................CGATCCTTGGGATTCTGCTC.......................................................................... | 20 | 1 | 3.00 | 3 | 2 | 0 | 0 | 1 |
| ......................................................................................................GGGTAA--ACTCCC----------------------------------------------------------------------------AAGGAT-----------------------C.................................................................................................................................................. | 19 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 |
| .....................................................................................................TGGGTAA--ACTCCC----------------------------------------------------------------------------AAGGAT-----------------------CAGC............................................................................................................................................... | 23 | 1 | 3.00 | 3 | 0 | 0 | 0 | 3 |
| .....................................................................................................TGGGTAA--ACTCCC----------------------------------------------------------------------------AAGGAT-----------------------................................................................................................................................................... | 19 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| ....................................................................................................CTGGGTAA--ACTCCC----------------------------------------------------------------------------AAGGAT-----------------------................................................................................................................................................... | 20 | 1 | 2.00 | 2 | 0 | 0 | 0 | 2 |
| ......................................................................................................GGGTAA--ACTCCC----------------------------------------------------------------------------AAGGAT-----------------------CAGGT.............................................................................................................................................. | 23 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| ......................................................................................................GGGTAA--ACTCCC----------------------------------------------------------------------------AAGGAT-----------------------CA................................................................................................................................................. | 20 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| .....................................................................................................TGGGTAA--ACTCCC----------------------------------------------------------------------------AAGGAT-----------------------CAGGG.............................................................................................................................................. | 24 | 1 | 2.00 | 2 | 1 | 0 | 0 | 1 |
| .....................................................................................................................................................................................................................................................................................TCCTTGGGATTCTGCTCTCG....................................................................... | 20 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| ....................................................................................................CTGGGTAA--ACTCCC----------------------------------------------------------------------------AAGGAT-----------------------CAG................................................................................................................................................ | 23 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................ATCCTTGGGATTCTGCTCTCGCTT.................................................................... | 24 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 |
| ..................................................................................................................................................................................................................................................................................CGATCCTTGGGATTCTGCT........................................................................... | 19 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................CCCGATCCTTGGGATTCTGCTCTCGG...................................................................... | 26 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 |
| .......................................................................................................GGTAA--ACTCCC----------------------------------------------------------------------------AAGGAT-----------------------CA................................................................................................................................................. | 19 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................ATCCTTGGGATTCTGCTCTCGGC..................................................................... | 23 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................CTGGGTAA--ACTCCC----------------------------------------------------------------------------AAGGAT-----------------------CAGG............................................................................................................................................... | 24 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 |
| ..................................................................................ATGTAAGCTGGCCTGTCCC........................................................................................................................................................................................................................................................................... | 19 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 |
| .....................................................................................................................................................................................................................................................................................TCCTTGGGATTCTGCTCTCGGTA.................................................................... | 23 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ......................................................................................................GGGTAA--ACTCCC----------------------------------------------------------------------------AAGGAT-----------------------CT................................................................................................................................................. | 20 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ......................................................................................................GGGTAA--ACTCCC----------------------------------------------------------------------------AAGGAT-----------------------CAGGA.............................................................................................................................................. | 23 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................ATCCTTGGGATTCTGCTCTA........................................................................ | 20 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................ATCCTTGGGATTCTGCTCTCGGTT.................................................................... | 24 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 |
| .....................................................................................................TGGGTAA--ACTCCC----------------------------------------------------------------------------AAGGAT-----------------------CAGGTTT............................................................................................................................................ | 26 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................TCCTTGGGATTCTGCTCTAAA...................................................................... | 21 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................ATCCTTGGGATTCTGCTCTCT....................................................................... | 21 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .......................................................................................................GGTAA--ACTCCC----------------------------------------------------------------------------AAGGAT-----------------------CAGG............................................................................................................................................... | 21 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................GATCCTTGGGATTCTGCTC.......................................................................... | 19 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................ATCCTTGGGATTCTGCTCTCGGG..................................................................... | 23 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................CGATCCTTGGGATTCTGCTCTCGA...................................................................... | 24 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 |
| .....................................................................................................TGGGTAA--ACTCCC----------------------------------------------------------------------------AAGGAT-----------------------CAT................................................................................................................................................ | 22 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 |
| ....................................................................................................................................................................................................................................................................................ATCCTTGGGATTCTGCTCTT........................................................................ | 20 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 |
| ....................................................................................................................................................................................................................................................................................ATCCTTGGGATTCTGCTCT......................................................................... | 19 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 |
| ..................................................................................................................................................................................................................................................................................CGATCCTTGGGATTCTGCTCTCGGTT.................................................................... | 26 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 |
| ....................................................................................................CTGGGTAA--ACTCCC----------------------------------------------------------------------------AAGGA........................................................................................................................................................................... | 19 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................ATCCTTGGGATTCTGCTCTCGC...................................................................... | 22 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................CTTGGGATTCTGCTCTCGG...................................................................... | 19 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................TCCTTGGGATTCTGCTCTCGGA..................................................................... | 22 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 |
| ..................................................................................................................................................................................................................................................................................CGATCCTTGGGATTCTGCTCGG........................................................................ | 22 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 |
| ....................................................................................................................................................................................................................................................................................ATCCTTGGGATTCTGCTTA......................................................................... | 19 | 6 | 0.17 | 1 | 0 | 0 | 0 | 1 |