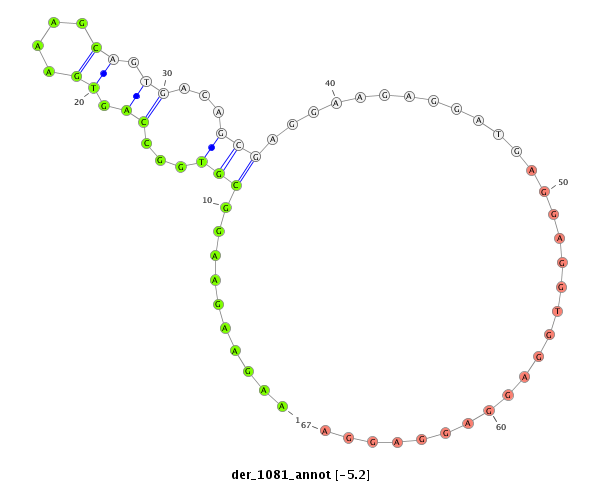
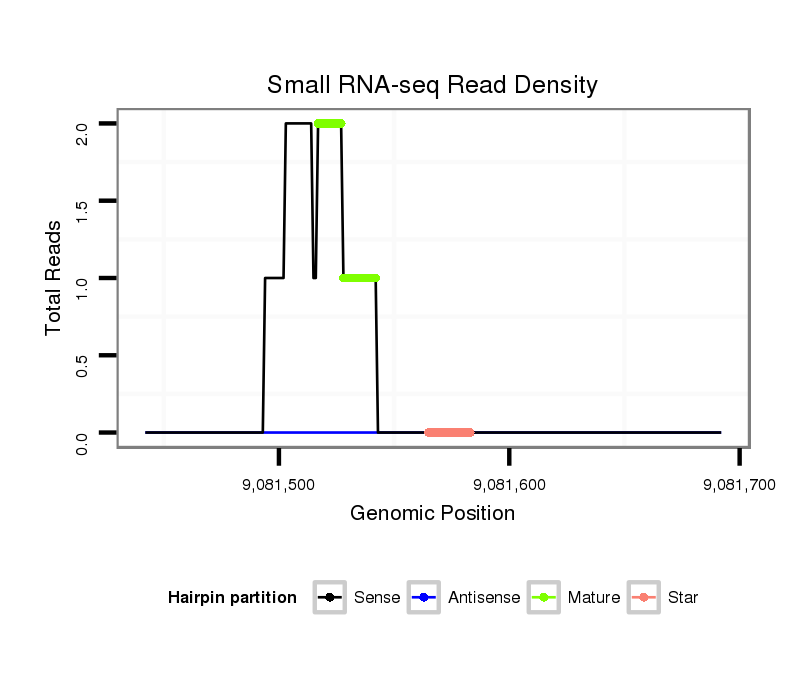
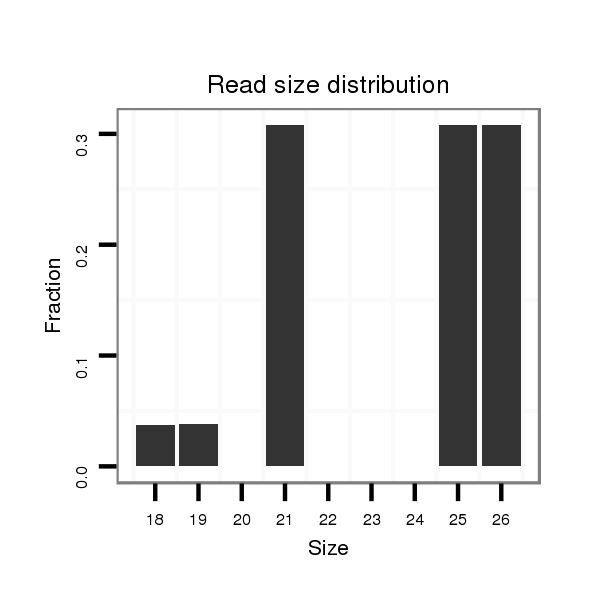
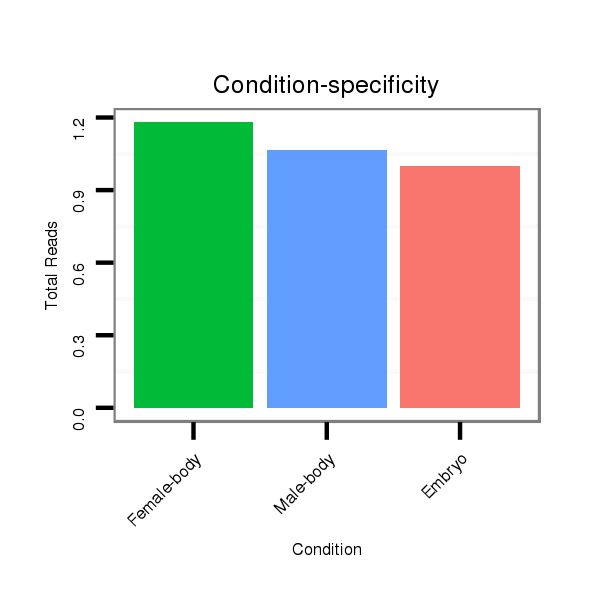
| droEre2 |
scaffold_4845:9081442-9081692 + |
der_1081 |
AG-GCG--------------GCGTCTCCAGTGCGTAATAGTGCGGCAACTACGACGCGAGCCGAGGTAA---GTG----TGAATG--------------------------AGGCAAATACA-------------------------GAAGAAGA------------------------------AGGCGTGGCCAGTGAAAGCAGTGACAGCGAGGAAGAGGATGAGGAGGT------GGAGGAG------------GAGGA--GGAAGAAGA---CGAAGA-GGAAGACGA----------GGA---TGAGGAGGAGGGGGAAGAACAAGCGGAGGAATCTGTA-------------------------------------------------------------------------------GAAGATACGGGTAGGGAA------------------CTGCTTAACCTCCC--AGAAGAAGC-TCTG |
| droYak3 |
2R:14307860-14308110 - |
dya_571 |
AG-GCG--------------GCGTCTCCAGTGCGTAGTAGTGCGGCATCCACGACGCGAGCCGAGGTAA---GTG----TGAATG--------------------------AGGCAAATACA-------------------------GAAGAAGA------------------------------AGGCGTGGCCAGTGAAAGCAGTGACAGCGATGAAGAGGAGGAGGATGT------AGAGGAG------------GAAGA--TGAAGAAGAAGACGAGGA-GGAGGAAGA----------TGA---TGAGGAGGAGGGTGAACAACAAGTA---GAACAAGTA-------------------------------------------------------------------------------GAAGAAACGGGTAGGGAA------------------TTGCTTAACCTCCC--GGAAGAGTC-ACTG |
| droSec2 |
scaffold_1:12392166-12392398 + |
|
AG-GCG--------------GCGTCCCCAGTGCGTAATAGTGCGACATCCACGACGCGAGCAGAGGTAA---GTG----TGAATG--------------------------AGGCAAATACA-------------------------GAAGAAGA------------------------------AAGCGTGGCCAGTGAAAGCAGTGACAGCGAAGAAGAGGAGGAGGAGGA---------------------AGAAGAAG---------------------AAGAAGAGGT----------GGA---GGAGGAGGAGGAGGAAGAACAAGTAGAGGAATCTGTA-------------------------------------------------------------------------------GAAGGGACTCCTAGGGAA------------------TTGCTTAACCTACC--AGAGGAAGC-TCTG |
| droSim2 |
2r:15534855-15535084 + |
dsi_25601 |
AG-GCG--------------GCGTCCCCAGTGCGTAATAGTGCGGCATCCACGACGCGAGCAGAGGTAA---GTG----TGAATG--------------------------AGGCAAATACA-------------------------GAAGAAGA------------------------------AGGCGTGGCCAGTGAAAGCAGTGACAGCGAAGAAGAGGAGGAGGAGGA---------------------AGAAGAAG---------------------AAGATGA-------------GGT---GGAGGAGGAGGAGGAAGAACAAGTAGAGGAATCTGTA-------------------------------------------------------------------------------GAAGGGACTCCTAGGGAA------------------TTGCTTAACCTACC--AGAGGAAGC-TCTG |
| dm3 |
chr2R:14885879-14886117 + |
|
AG-GCG--------------GCGTCCCCAGTGCGTAATAGTGCGGCATCCACGACGCGAGCTGAGGTAA---GTG----TGAATG--------------------------AGGCAAATACA-------------------------GAAGAAGA------------------------------AGGCGTGGCCAGTGAAAGCAGTGACAGCGAAGAAGATGAGGAAGTTGA---------------------AGATGAAGA--TGAAGA-------------AGAAGA-------------GGT------AGACGAGGAGGAGGAACAAGTAGAAGAATCTGTAGAAGAA-------------------------------------------------------------------------GAAGAGGCTCCCAGGGAA------------------TTGCTGAACCTACC--AGAGGAAGC-TCTG |
| droEug1 |
scf7180000409663:443316-443530 - |
|
AG-GCA--------------GCGTCTCCAGTGCGTAATAGTGCCGCAGCTACGACGCGAGCCGAGGTAA---GTG----TGAATG--------------------------AGGCAAATACA-------------------------GAAGAAGA------------------------------AGGCGTGGCCAGTGAAAGCAGTGACAGCGAAGAAGAGGAGGAGGAAGA---------------------AGAAGAGG----------------------------------------------------ATGAGGGTCCAGAACAAGCAGAAGAATTTGCA-------------------------------------------------------------------------------GAAGTGGCTTCTAGGGAC------------------CGGCTTAACCCACC--GGAGGAATC-CCTG |
| droBia1 |
scf7180000302292:1478900-1479102 - |
|
AG-GCG--------------GCGTCTCCAGTGCGTAACAGTGCAGCCTCTACGACGCGAGCCGAGGTAA---GTG----TGAATG--------------------------AGGCAAATACA-------------------------GAAGAAGA------------------------------AGGCGTGGCCAGTGAAAGCAGTGACAGCGAGGAAGAGGAGGAGGAGGA---------------------G-------------------------------------------------------------GCGGGGGGAGAGCAACCGGAAGAATCTGTT-------------------------------------------------------------------------------CAAGTGGTTCTACAGGAT------------------GTGCTTAGTCT-----AGGAGAGCC-TCAG |
| droTak1 |
scf7180000415991:355335-355564 + |
|
AG-GCG--------------GCGTCTCCAGTGCGTAACAGTGCGGCCTCAACGACGCGAGCCGAGGTAA---GTG----TGAATG--------------------------AGGCAAATACA-------------------------GAAGAAGA------------------------------AGGCGTGGCCAGTGAGAGCAGTGACAGCGAAGAAGAGGAGGAGGAGGA---------------------GGATGAGGA---------------------GGAAGAATC----------GGT------AGAGGAGGGGGCAGAACAAGCTGAAGAATCTGTA-------------------------------------------------------------------------------GAAGGGGCTCCTAGGGAA------------------TTGCTTAATCCCCC--AGAAGAATC-TCTG |
| droEle1 |
scf7180000491228:23867-24075 - |
|
AG-GCG--------------GCGTCTCCAGTGCGTAATAGTGCGGCATCTGCGACGCGGGCTGAGGTAA---GTG----TGAATG--------------------------AGGCAAATACA-------------------------GAAGAAGA------------------------------AGGCGTGGCCAGTGACTGCAGTGACGGCGAAGAGGAAGAAGAAGAGGA---------------------A----------------------------------------------------------GAGGAGGGG------------GAAGAATCTGTAGAGGC-------------------------------GGA---------------------------AATC---------GAAGAGGCTCCCAGGGAA------------------TCTCTTAACCTTCC--AGAGGAATC-TCTG |
| droRho1 |
scf7180000779618:14868-15073 + |
|
AG-GCA--------------GCGTCTCCAGTGCGTAATAGTGCGGCGACTACGACGCGAGCTGAGGTAA---GTG----TGAATG--------------------------AGGCAAATACA-------------------------GAAGAAGA------------------------------AGGCGTGGCCAGTGACAGCAGTGACAGCGAAGAAGAGGGGGAGGAGGA---------------------GGAGGAGG-------------------------------------------------------------ATGAGGAGGGGGAGGAATCCGTA-------------------------------------------------------------------------------CAGGAAGCTACCGGAAAT------------------GTGCTTAACCTTCC--AGAAGAAGC-TCTT |
| droFic1 |
scf7180000454044:681142-681425 + |
|
AG-GCG--------------GCGTCTCCAGTGCGTAACAGTGCCGCATCGACGACGCGAGCCGAGGTAA---GTG----TGAATG--------------------------AGGCAAATACA-------------------------GAAGAAGA------------------------------AGGCGTGGCCAATGACAGCAGTGACAGCGAAGAAGAAGTTGAAGAGGA------AGGGGAG------------GAGGA--GGAGGAGGA---GGAGGA-GGAGGA-------------GGA---GGAGGAGGAGGAGGA---------AGAAGAATCAGCAGGGGAAGGGGTAGAACAGGTGG-AGGAACCTTCAGTGGG---------TGT---------------GGTA---------GAAGAGGCTTCAAGAGAA------------------GTTCTTAATCTACC--AGAAAAGGC-TTCG |
| droKik1 |
scf7180000302682:642209-642429 + |
|
AG-GCG--------------GCGTCTCCAGTGCGTAATTCGGCGGCAGCGGCAACGCGAGCTGAGGTAA---GTG----TGAATG--------------------------AGGCAAATACA-------------------------GAAGAAGA------------------------------AGGCGTGGCCAGTGACAGCAGTGACAGCGAGGAAGAGGAGGAGGATGA------TGAGACT------------GAGGA----------------------------------------GGAAG---------------ATGATGAAG------------TA-------------------------------------------------------------------------------GAAGAGTCTCCTGAGGTCCTGCCAGAGTTGGCCCAAGGTCTTAGCCCCCC--AGAAGTGCC-TGCA |
| droAna3 |
scaffold_13266:17449138-17449346 - |
dan_2079 |
AG-GCG--------------GCGTCTCCAGTGCGTAACAGTGCAGCAACCGCGACGCGGGCCGAGGTAA---GTG----TGAATG--------------------------AGACAAATACA-------------------------GAAGAAGA------------------------------AGCCCTGGCCAGCGAAGGCAGTGATAGCGAAGAGGAGGAGGAAGAAAC----------------------------------------------------------------------------------------------------AGAAGAGTCTGAAGAAAGTGAAGTAGAACAGGAGGC----A---------------------------------------------------GGTGTGGCGCCGCAGGAC------------------ACGCTTAATGTTCC--AGA---AGC-TAGC |
| droBip1 |
scf7180000396735:710723-710935 - |
|
AG-GCG--------------GCGTCTCCAGTGCGTAACAGTGCAGCAACCGCGACGCGGGCCGAGGTAA---GTG----TGAATG--------------------------AGACAAATACA-------------------------GAAGAAGA------------------------------AGCCATAGCCAGCGAAAGCAGTGACAGCGAAGAGGAGGAGGAAGAAAC----------------------------------------------------------------------------------------------------AGAAGAGTCTGAAGAAAGTGAAGTAGAACAGGAGGC----A---------------------------------------------------GAAGTAAGGCCACAGGAC------------------CAGCTTGATGTGCC--AGAAGCTAGCCAGG |
| dp5 |
3:5960643-5960838 + |
|
AG-GCG--------------TCATCTCCAGTGCGTA---CAGCGACAGCAGCGACGCGAGCCGAGGTAA---GTG----TGAATG--------------------------AGGCAAATACA-------------------------GAAGAAGA------------------------------AGGCGTGGCCAGTG---GCAGTGACAGCGAGGTAGACGAGGAGAATGA---------------------GGAACAGGGGG------------------CAGAAGA-------------GGAAGATCAGCAGCAGCAG---------------------------------------------GC----AGAGGTAGAGGA---------------------------AGTCGAAAAC--------GACTCCGAAGA-------------------------------------------------- |
| droPer2 |
scaffold_2:6161599-6161794 + |
|
AG-GCG--------------TCATCTCCAGTGCGTA---CAGCGACAGCAGCGACGCGAGCCGAGGTAA---GTG----TGAATG--------------------------AGGCAAATACA-------------------------GAAGAAGA------------------------------AGGCGTGGCCAGTG---GCAGTGACAGCGAGGTAGACGAGGAGAATGA---------------------GGAACAGGGGG------------------CAGAAGA-------------GGAAGATCAGCAGCAGCAG---------------------------------------------GC----AGAAGTAGAGGA---------------------------AGTCGAAAAC--------GACTCCGAAGA-------------------------------------------------- |
| droWil2 |
scf2_1100000004510:2489661-2489862 + |
dwi_441 |
AG-GCA--------------GCATCTCCAGTGCGTA---CAGCCACGACAAGCGCGCGAGCCGAGGTAA---GTG----TGAATG--------------------------AGGCAAAT----------------------------GAAGAAGA------------------------------AGATCTGTCCAGTGA---CAGTGGCAGCGAGGAAGAGGACGACGAAGA---------------------GGAAGATGATG------------------CAG-------CAGTTGAACAGAAAGAAGTAGTGCCACAG---------------------------------------------GT----ACAAGAGGAGGA---------------------------AAAGGAAAATGAGGATGAAGCTTC------------------------------------------------------- |
| droVir3 |
scaffold_12875:19589786-19590036 - |
dvi_7992 |
ATCAGTCGGCGACAGCGGCGGCATCTCCAGTGCGTACAACGACAGCGTCGACAGCGCGAGCCGAGGTAA---GTG----TGAATG--------------------------AAACAAAT----------------------------GAAGAAGA------------------------------AGGCGTGGCCAGTGACAACAGTGGCAGCAGCGAGACGGAGAGTGAGACGGGAAGTGAGAGC---------------GAGG------------------CGGAGGA-------------AGAAGAGCAGCAGCAGCAG----CAGCAGCAGGCGCAGTTGGC--------------------GC-AGGCACCCGCTGAGGA---------TCA---------------AGTGGATAACGA--TTCCAGCTCCG----------------------------------------------------AG |
| droMoj3 |
scaffold_6496:6639868-6640151 - |
|
AGAGCA------G-------AGCATGCCAGTGATAAATATTGTGCCGCCAGAG-AATGAGCTGATGGAGAAGGCGTCAGAGAATGAATGCAATATGGATGTCATAAGCGATAGGCATATAGAGCAGGAGATTGAGGAGAAGCAGGAGGAGGAAGA------------------------------GGGCGA------------------TGGCGAGGAAGTTAAAGTGGAAGT------AGCAGAGGAGGAAGAGGAAGAGATTA------------------CAGATGATGA----------GGA---GGAGGAGGAGGAGGAAGATGAAGAAAATGAGATTGAG-------------------------------------------------------------------------------G--------CAGAGGTGA------------------AGGCAGAGCCAATTGCAGA--AGCA-GCTG |
| droGri2 |
scaffold_15245:17961626-17961895 + |
|
AA-GCG-----ACAACAGCATCATCTTCAGTGCGTACAGCAACAACGGCGACAGCACGAGCCGAGGTAA---GTG----TGAATG--------------------------AAACAAATGAA-------------------------GAAGAAGAAGTGGCAGCAGCAGCAGAAAATGATGAAGTAGGTGTGGCCAGTGACGACAGTGGCAGCAGCGACATTGAGAGCGAAAC---------------------AGAACAAG---------------------AGCGTGAAC-AAGTTCAAGAGCAAGAGTTGCAACCACAA---------------------------------------------GC----AGTAGCGGTGATAAATGAAGATCAAGTGGACAACGACGACGTCG---------ATG--ACGCC------------------------------------------------------T |