ID:dan_968 |
Coordinate:scaffold_12943:4659745-4659895 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
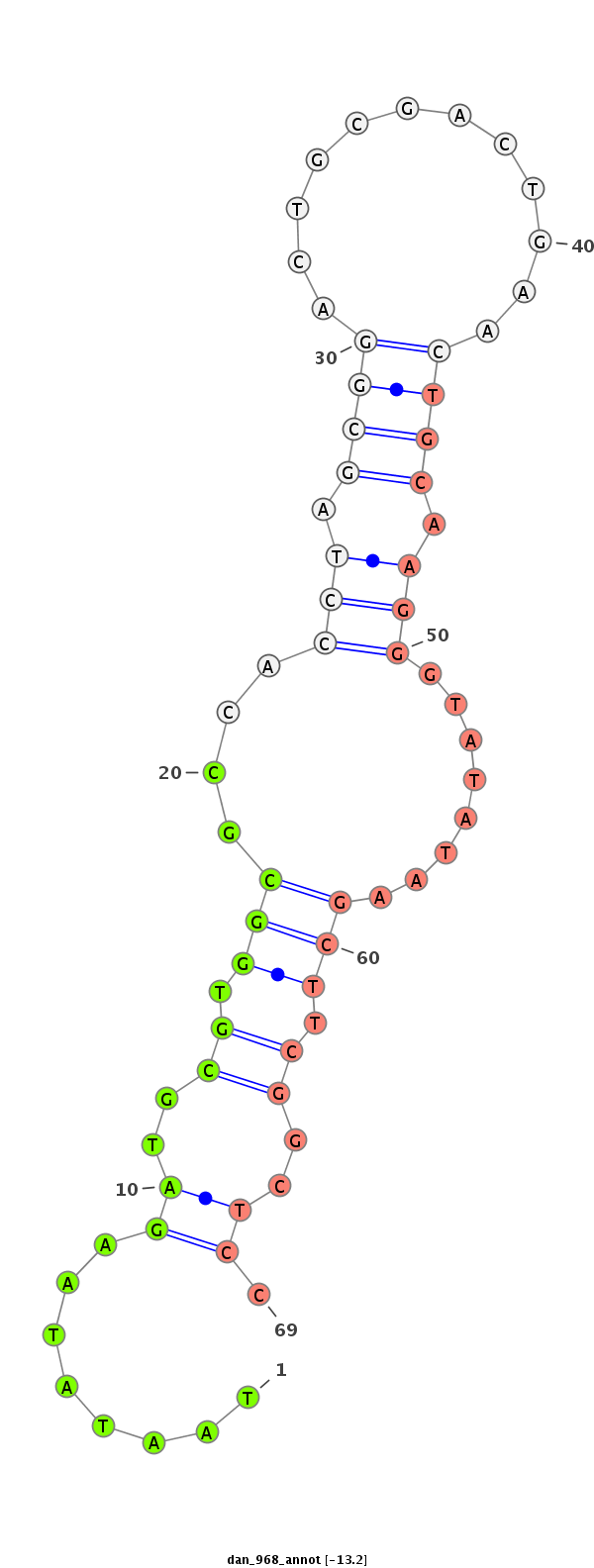
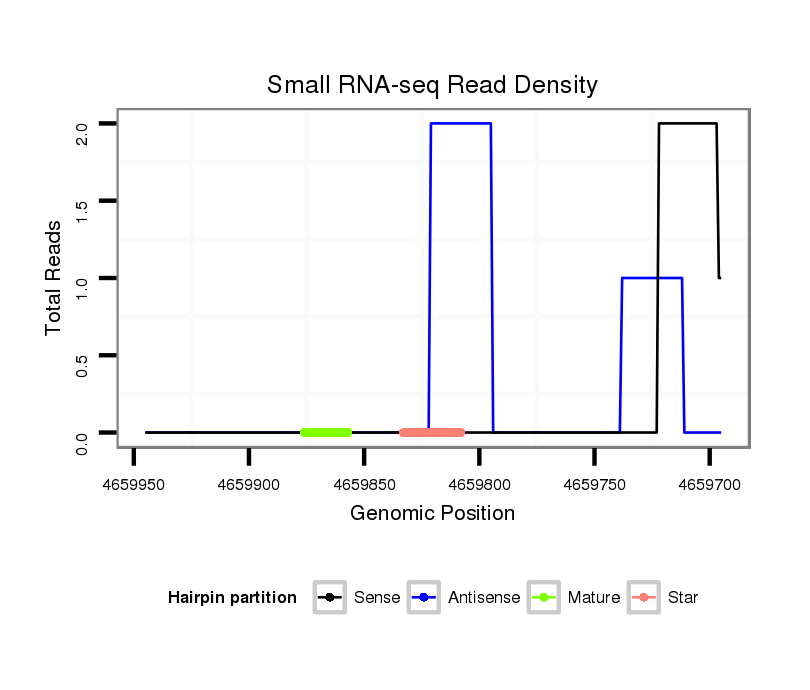
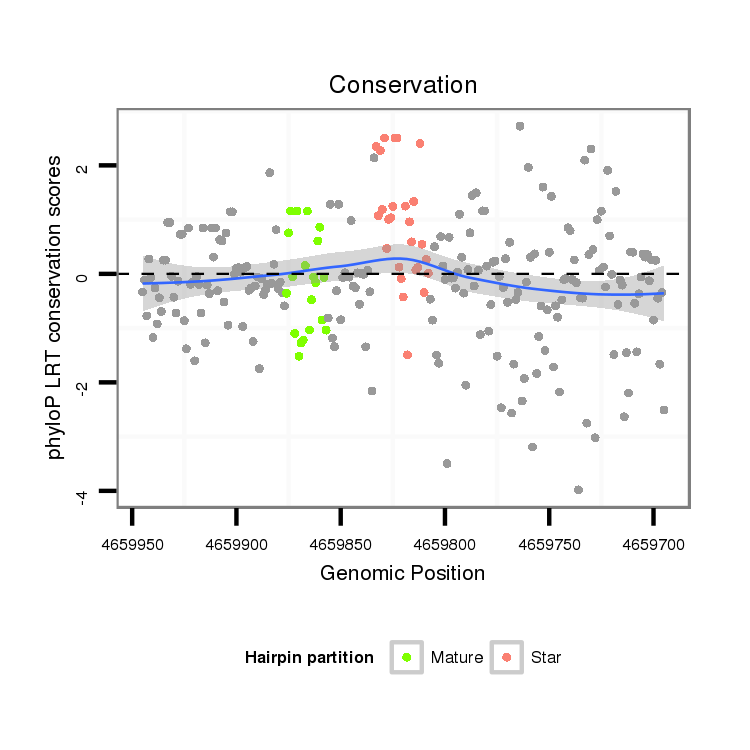
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -12.8 |
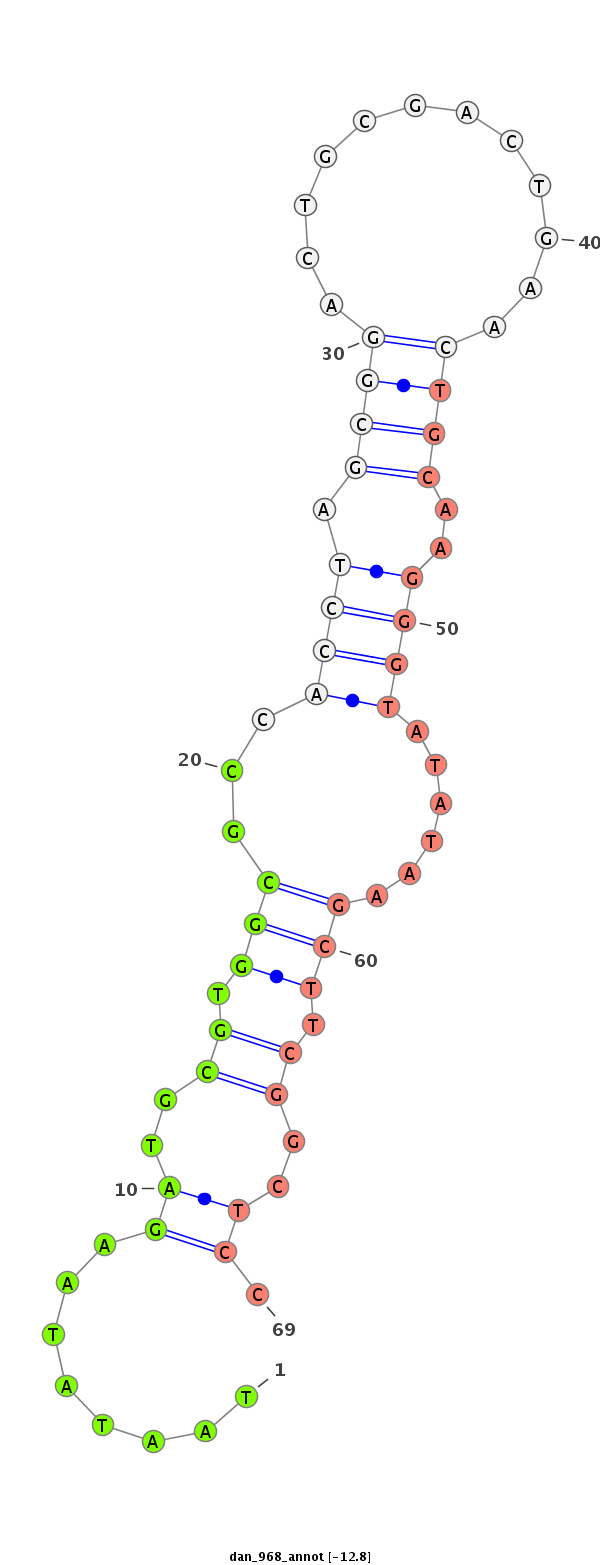 |
CDS [Dana\GF20672-cds]; exon [dana_GLEANR_379:2]; intron [Dana\GF20672-in]
| Name | Class | Family | Strand |
| Hoana8 | DNA | hAT-hobo | - |
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TAGATATTTAAAAAACCTTTTCAACGATGTAGGTTTAACGACTATAACCAGACTATAAACGATCCGCTATAATATAAGATGCGTGGCGCCACCTAGCGGACTGCGACTGAACTGCAAGGGTATATAAGCTTCGGCTCCGCCTGAAGTTAGCTTTCCTTTCTTGTTTTATTATTTCCTTTTCTTGTTTTACTAAAATTTCAGAAAATTTTCAAAAAAACAATTATTCAGCGGGTAGCCCCTAAAAGGCGGGT *********************************************************************........((..((.(((....(((.((((............)))).)))........))).))..)).***************************************************************************************************************** |
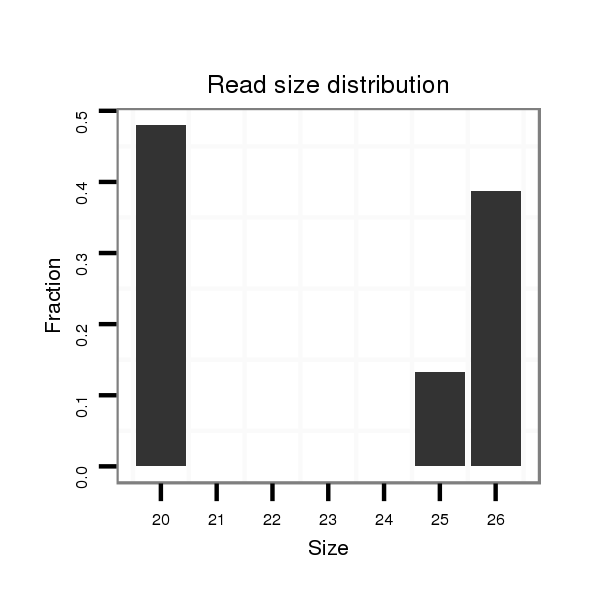
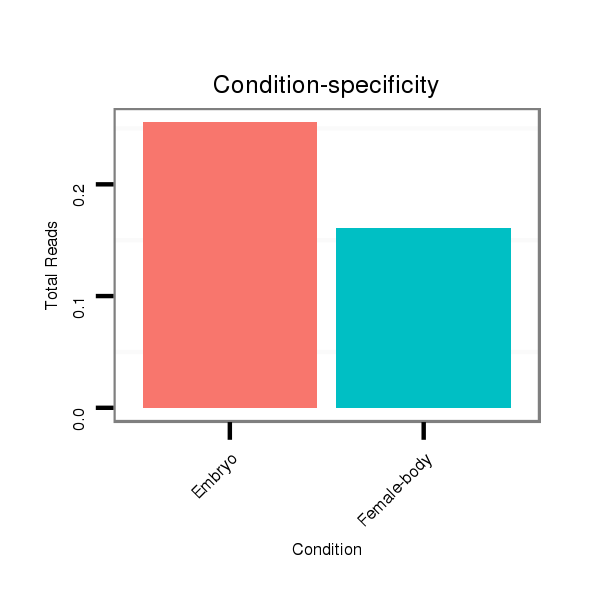
Read size | # Mismatch | Hit Count | Total Norm | Total | M044 female body |
M058 embryo |
V039 embryo |
|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................................................................................................................TTCAGCGGGTAGCCCCTAAAAGGCGGGT | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ...............................................................................................................................................................................................................................TTCAGCGGGTAGCCCCTAAAAGGCGG.. | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ...............................................................................................................................................................................................................................TTCAGCGGGTAGCCCCTAAACGGCGG.. | 26 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| .....................................................................TAATATAAGATGCGTGGCGC.................................................................................................................................................................. | 20 | 0 | 20 | 0.20 | 4 | 0 | 0 | 4 |
| .................................................................GATCTAATATAAGATGCGAGGCGCC................................................................................................................................................................. | 25 | 3 | 9 | 0.11 | 1 | 0 | 1 | 0 |
| ................................................................................................................TGCAAGGGTATATAAGCTTCGGCTCC................................................................................................................. | 26 | 0 | 18 | 0.11 | 2 | 1 | 1 | 0 |
| .................................................................................................................................................GTTAGCTTTCCTTTCTTGTTTTATTACTT............................................................................. | 29 | 1 | 15 | 0.07 | 1 | 0 | 1 | 0 |
| ..................................................................................................................CAAGGGTATATAAGCTTCGGCTCCG................................................................................................................ | 25 | 0 | 18 | 0.06 | 1 | 1 | 0 | 0 |
| ...........................................ATAACCAGACTAGGAAGGA............................................................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 |
| ................................................................................................................................................AGTTAGCTTTCCTTTCTTGTTTTATT................................................................................. | 26 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
|
ATCTATAAATTTTTTGGAAAAGTTGCTACATCCAAATTGCTGATATTGGTCTGATATTTGCTAGGCGATATTATATTCTACGCACCGCGGTGGATCGCCTGACGCTGACTTGACGTTCCCATATATTCGAAGCCGAGGCGGACTTCAATCGAAAGGAAAGAACAAAATAATAAAGGAAAAGAACAAAATGATTTTAAAGTCTTTTAAAAGTTTTTTTGTTAATAAGTCGCCCATCGGGGATTTTCCGCCCA
*****************************************************************************************************************........((..((.(((....(((.((((............)))).)))........))).))..)).********************************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | M044 female body |
M058 embryo |
V106 head |
V055 head |
V039 embryo |
V105 male body |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................ATTCGAAGCCGAGGCGGACTTCAATCG.................................................................................................... | 27 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................AAGTTTTTATGTTAATAAG......................... | 19 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................AAGTTTTTTTGTTAATAAGTCGCCCAT................. | 27 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................GGTGGATCGCCTGACGCTGAC.............................................................................................................................................. | 21 | 0 | 20 | 0.80 | 16 | 0 | 0 | 16 | 0 | 0 | 0 |
| ..........................................................................................TGGATCGCCTGACGCTGACTTGACG........................................................................................................................................ | 25 | 0 | 20 | 0.50 | 10 | 0 | 0 | 10 | 0 | 0 | 0 |
| ................................................................................CGCACCGCGGTGGATCGCCTGACGCT................................................................................................................................................. | 26 | 0 | 20 | 0.35 | 7 | 0 | 7 | 0 | 0 | 0 | 0 |
| ...........................................................................................GGATCGCCTGACGCTGACTT............................................................................................................................................ | 20 | 0 | 20 | 0.35 | 7 | 7 | 0 | 0 | 0 | 0 | 0 |
| ......................................................TATTTGCTAGGCGATATTATATTCT............................................................................................................................................................................ | 25 | 0 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................TTTGCTAGGCGATATTATATTCT............................................................................................................................................................................ | 23 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................ACTTGACACTCCCATAGATTCG.......................................................................................................................... | 22 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .........................................................................................GTGGATCGCCTGACGCTGACTTGACG........................................................................................................................................ | 26 | 0 | 20 | 0.20 | 4 | 4 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................GACGCTGACTTGACGTTCCCATATAT............................................................................................................................. | 26 | 0 | 20 | 0.20 | 4 | 4 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................AAAGAAAAAAATAAGAAAGGAAAA....................................................................... | 24 | 2 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................CAATCGAAAGGAAAGAACAAAATAAT................................................................................ | 26 | 0 | 20 | 0.20 | 4 | 0 | 4 | 0 | 0 | 0 | 0 |
| .........................................................................................GTGGATCGCCTGACGCTGACTTGACGTT...................................................................................................................................... | 28 | 0 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................CTACGCACCGCGGTGGATCGCCTGAC.................................................................................................................................................... | 26 | 0 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................AGAACGAAATGATTTTGAA..................................................... | 19 | 2 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................GGATCGCCTGACGCTGACTTGAC......................................................................................................................................... | 23 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................AAGGAGAAGAACAAAGTGA............................................................ | 19 | 2 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| .....................................................................................................................................................................AATAATAAAGGAAAAAGACAAAA............................................................... | 23 | 2 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................................................................................ACAAGAAAGGAAAAGAACAA................................................................. | 20 | 2 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................CGAACGAAATGATTTTAA...................................................... | 18 | 2 | 14 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................AAGAAAAAGATCAAAATGGTTT......................................................... | 22 | 3 | 18 | 0.06 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................................................GTTCCCATATATTCGAAGCCGAGGC................................................................................................................ | 25 | 0 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................TCAATCGAAAGGAAAGAACAAAATAAT................................................................................ | 27 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................CCTGACGCTGACTTGACGTTCCC................................................................................................................................... | 23 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................AAGCCGAGGCGGACTTCAATCGAAAG................................................................................................ | 26 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................TGCTTGGCGATATGATATC.............................................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ....................................................................................................................................CCGAGGCGGACTTCAATCGAAAGG............................................................................................... | 24 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................TCGAAAGGAAAGAACAAAATAAT................................................................................ | 23 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................TGACTTGACGTTCCCATATAT............................................................................................................................. | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................AGCCGAGGCGGACTTCAATCGAAAG................................................................................................ | 25 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................AGACAGGAAAAGAACAAA................................................................ | 18 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................AGAACAAAATAAAGAAGGA.......................................................................... | 19 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................AACAAAGGAAGAGAACAAAA............................................................... | 20 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................ATCGCCTGACGCTGACTTGACGTTCCC................................................................................................................................... | 27 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................ATCGCCTGACGCTGACTTGACGTT...................................................................................................................................... | 24 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................CTTCAATCGAAAGGAAAGAACAAAATAA................................................................................. | 28 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droAna3 | scaffold_12943:4659695-4659945 - | dan_968 | TAGATATTTAAAAAACCT--TTT-CAACGATGTAGG------------------TTT-------------------AACG---------------ACT--ATAACCAG--ACTAT-AAA-C-GATCCGC----TATAA-TATAAGATG--CG---------------------------------------------------------------------------------------------------TGGCGC--CACCTAGCGGACTGCGA-------CT-------G----------------------AACTGCAAGGGTATATAAGCTTCGGCTCC-GCCT-G-AAGTTAGCTTTCC------TTTCTTGTTTTA-----TTATTTCCTTTTCT-TGT---------------------TT----------------------------TA------------------CTAAAATT--------------------------TC-----------AGAAAATTTT----------------------------------------CAAAAAAACA-------ATT------ATTCAGCGGGTAGCCCCTAAAAGGCGGG-T |
| droBip1 | scf7180000396641:89329-89553 + | TAGATTTTTTA---------TTG-TAATTAA-TTGG------------------TTTTATTATGACGTAATCATAA----G-----------------------------------------GATCCGCCAACT---AT-ATCCGATG-----------------------------------------------------------------------------------------------------TTT-GTGA--TATATATC-------------------CGGTTTT----------------------AGCTGCAAGGGTATATTAACTTCGGTTCCGCCCG---AAGTTAGCTTTCT------TTTCTTG-----TTT-----TTTCTGCCGTTGCTT----------------------T------------------------------------------------------------------TGGCTTTAAAAAGC---------------ATAAAA---------------------------TAATAAAA----------CAATAAAAA---AAC-A-----GAAA-----------G--ACTAGACAGCGGG-T | |
| droKik1 | scf7180000301787:6183-6406 - | TAGATGTATACAAAA----------------------------------------------------------------------------------------------TGTAAAGC--TGGGA-----ATGCA---AAACT---GTAACTGTCAAACTGTAAACATAATAAGTA--------------------------------------------------------------------------TAG-GTAA--AATGTAAT-----------------G-AAATAAT----------------------ATCTGCAAGGGTATACAAACTTCGGCGT--GCCG---AAGTTAGCT-TCC------TTTCTTG-----TTT-----TTATATCAT-TTCGC---------------------TG-----------------GTTTT-----------------------------------------------------------------------------T------AAACTGACAAATATTAGAAAATT----------------AGTAAAAATAATA----TTT------TTTCGGTGGCAAGTTTC------------T | |
| droFic1 | scf7180000454058:29426-29724 + | TATATATTA--------------A--------------------------------------------------------ATT------T-------------------TATCAAAA--TCGGA-----CGACT---AT-ATCATATAGCTGTCATATGAACAATATGGTAAATAATAGAAAAATCGTCATAGATTCGATGTTATT--------------------------TTCCTTTATGTATTTTGTT----------------------TGA-------CATATTTAAT----------------------GACTGCAAGGGTATATAAGCTTCGGCTT--GCCG---AAGCTAGCT-TCC------TTTCTTG-----TTA-----GC--------TTAAATTTGGCACTATTGCAATGTATTTT-----------TTTTCTGCACAAC---GCAATATCCGGTTAAATTTTACAATTTTCTAGAAAAATTTCGTTCGTATA-------------------------------------------------------------------------------------------------AGCGGTTTGTCGCTAAAAG------- | |
| droEle1 | scf7180000490204:23182-23401 + | TAGGAACTC--------------A--------------------------------------------------------AAT------------------------A--ATTCAGCTG-T-AAA-----------------------------------------------------------TCATCATAGCTTCGAGGCTTTTAAACATACACGCAAGTAAATCATAATTTCAATTAAA--TGTTCTT----------------------TAA-------TTTTTGCAAT----------------------TGCTGCAAGGGTATATGAACTTCGGTTT--GCCG---AAGTTTGAT-TCC------TTTCTTG-----TTT-----TTATTTTTTTT-CAT---------------------TC-----------------ATCTT-----------------------------------------------------------------------------T------GCATCTGC--ATATTCACAAATT----------------AAAAAAAAAAATT-------------TTTT-------------------------A | |
| droRho1 | scf7180000780120:81798-82000 + | TGAGTATTA--------------A--------------------------------------------------------GAATGCATCATATGA------------------AT-AA--CCGATCC----------------------------------------------------------------------------------------------------------------------------------------------------------------GGTTTTGGAGAATATAATTTACAGAATATTCTGCAAGGGTATATCAACTTCGGCTCCGTCCG---AAGTTAGCT-TTC------TTTCTTG-----TTTTATAC-----TTAC-T-----TA------------------------------------------------TA------------------CTT-------------------------------AC-----------TTTTAATATCGTAACT-----------------TAGTAAAAACGAATTTTCAAAGAAA-GATA-------------TTTTAGC----AATTCCTAAAAG------- | |
| droBia1 | scf7180000302521:94015-94228 + | AAAATGTATGAAAAACAATG---AAATCATTTTTTC------------------T-T-------------------------------------TAAT--ATAACTAA--ATTCAGCAA-C-AATCCTT----AAAAA-TTTCACATG----------------------------------------------------GT--------------GTA---------------------AC--TAAAGTT----------------------TAT-------TATTTCTTAT----------------------AACTGCAAGGGTATACAAACTTCGGCTT--GGCC---AAGTTAACT-CCC------TTTCTTG-----TTT-----TTAATGAAC-TTTGCTTT----------------------TATTTATTTA-----------------A------------------TTT-------------------------------TC-----------TTTTAATTTTGAGACT----------------------------------------AACA-------A-------------------------------------T | |
| droTak1 | scf7180000415606:8429-8691 + | CAAATATGAGAAAATTCA--TTG-TAACTTTGTAGGAGGTAGGCGTTTTAATTCT-T-------------------GACT---------------ACTTAAAAACAAAATTCAAAAA--TAGAA-----ACGCTG-AA-TCTGGAATC--CC---------------------------------------------------------------------------------------------------TGGAACTGCACCGAGTGAGCATTAA-------TT-------T----------------------ATCTGCAAGGGTATATAAGCTTCGGCTG--GCCG---AAGGTAACT-TCC------TTTCTTGTTTAA-----TTG-----TATT-T---------------------------T-----------TTGTCTACTG------AA------------------AAAACGT---------------------------AT---------GCAAATAATCAT----------------------------------------GTTATGAGT---------------------GGAGCGTGGTCTATAAAGGACATA-T | |
| droEug1 | scf7180000408236:4878-4962 + | TAG--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CG-TTT------CCTCTTG-----TTT-----TATTATAGC-TTTGCTTT----------------------TATTTATTGA-----------------A------------------TCT-------------------------------AC-----------TCTTAATTTTGAGACT----------------------------------------AACA-------ATAAGTTG------------------------ACGAA-T | |
| dm3 | chr3R:765701-765758 + | TGT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACTGCAAAGGTATATATGCTTCGGCAT--GCCG---AAGCTATCT-TTT------TTTCTAT-----TTT-----TA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droSim2 | x:14966218-14966301 + | TTT-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTATTTCA-----TTA-----TTTT-T---------------------------T-----------------TCACAATTTCTA------------------CCAATATT--------------------------CC-----------AAAAAATATT----------------------------------------AAAATGAGTA-------A--------------CGG---GTATCTGATAGTCGGG-T | |
| droSec2 | scaffold_1:5681320-5681386 + | A----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTGCAAGGGCATACAAACTTCGGCTT--GTCG---CAGTTAATT-TCC------CTTCCTG-----TTA-----GC--------TTTGTTCT----------------------TG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CC | |
| droYak3 | 3R:984477-984570 + | A----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTGCAAGGGTATATAAGCGTCGGCTT--GTCG---AAGCTAGCA-TTA------TTTTTATTTAAG-----TTG------------------------------------------------------------------CA------------------TTAAAA-------------------------------------GGG----TTATATC----------------------------------------CAAAAAAAGA-------------GTAATT-------------------------CA | |
| droEre2 | scaffold_4929:175555-175672 + | A----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTACAAGGGTATATAAGCTTCAGCTT--GTCA---AACGCAGCT-TTC------TTTCTTG-----TTG-----GA--------TTAACGTGTGCC------------------GATTTATGTC-----------------G------------------TTT-------------------------------GCTCTTTATTCA----------TTGGATCCAC--A--------------------------------GA----ACT---------TTT----T--------------------CAC-AA | |
| dp5 | 3:13007579-13007690 - | -----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTGCAAGGGTATCTGAGGCATCGCTT--TCCCAAGATGCAGGTT-TTT------CTTCTTCTTTAG-----CTGCTTTCTGTTTT-GCT---------------------TT----------------------------TA------------------TTAATTT---------------------------TC---------G-------TTG---------------------------------------T-CAT---------------------------GGCAGACAGTCTTCAA---------A | |
| droPer2 | scaffold_3:556993-557139 + | TAATCCGCA--------------A--------------------------------------------------------CA-----GTAAATGA------------------AA-AA--CGGA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCCT--GTCT-G-TATATATCGTATG------ATTCTGT-----TTT-----TGG--TATC-T---------------------------TTTATTAGTT-----------------GCA------------------TTAATTCT--------------------------TA-----------GGAAAATTTT----------------------------------------TAAAAAAATA-TGGTCAGTT------GTATAGGCAGTCGTCTAGAAAAGCT---TG | |
| droWil2 | scf2_1100000004909:282048-282241 - | TAGAGAGTA--------------A--------------------------------------------------------AATTGTTTTGTTTGT-AT--ATTGATGA--ATTGAGTTG-C-AAA---------------------------------------------------------------------------------AAACAAACATGTAA---AA-------------------------T----------------------TAT-------TATTACCAAG----------------------GACTGCAAGGGTATATAAACTTCGGCATA-GCCG---ATGTTAGCT-TCT------TTAATAT-----TTT-----TC---TTTTCT-TTT---------------------TT-----GAGTG-----------------GGA------------------AAAAATTT--------------------------AA-----------TTAAGGATT---------------------------------------T-TATAAAAAAAATT-------------TTTTAG-----------------------C | |
| droVir3 | scaffold_12958:3107544-3107654 - | -----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTGCAAGGGTATTAGATCTTCGGCGT--GCCG---AAGATAGCCTTTC------TTTCTTG-----TTT-----TATTTTCGTCTTATT----------------------T------------------------------------------------------------------TGTGTTCATATAAGAGAG-----------TTTTAATTTA-----------------------------------------GAATAAATA---ATG-A-------------------------------------T | |
| droMoj3 | scaffold_6473:801941-802030 - | T------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCGAATATATCTGATCTTGTGCTT--ATTT---ATTTTCGTT-GTT------TGTTTTT-----TTT-----TTTTTTTTTTT-TTT---------------------TT-----------------TTCTC-----------------------------------------------------------------------------A------ATATCTAT--A---------------------------------------------------TTGTTT-------------------------TC | |
| droGri2 | scaffold_14853:7989917-7990051 - | ATGATGCAAGAG-------------------------------------------------------------------------------------------------TGCAGGGC--------T--------------------------------------------------------------------------------------------------------------------------------TGC--CAGCCAGCAGGCAGCCACAAGTTCTT--GAGTTGAACGAATATATTTTTTAAAA-----------------------TAGGCTT--TGAA---ATTTTTGTT-TTGTTTTTTTTTTTTG-----TTT-----TTTTTTTTTTT-TGT---------------------TT----------------------------TA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------TT |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/17/2015 at 11:50 PM