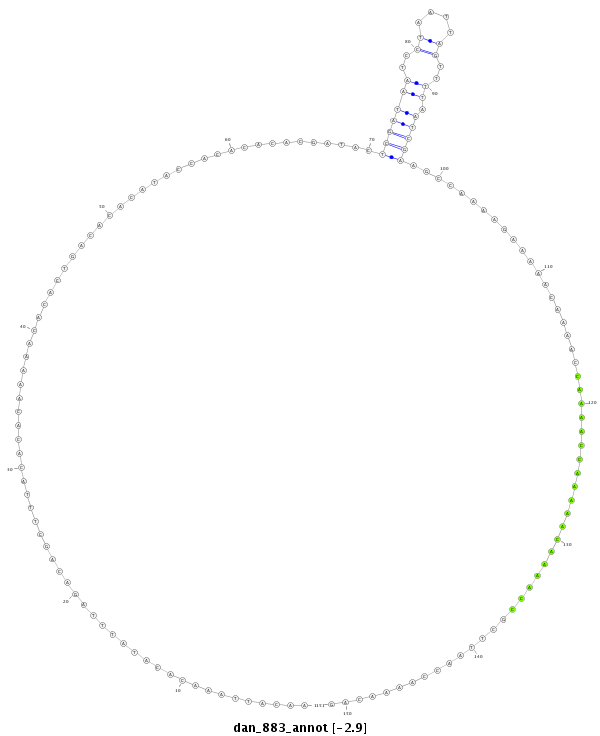
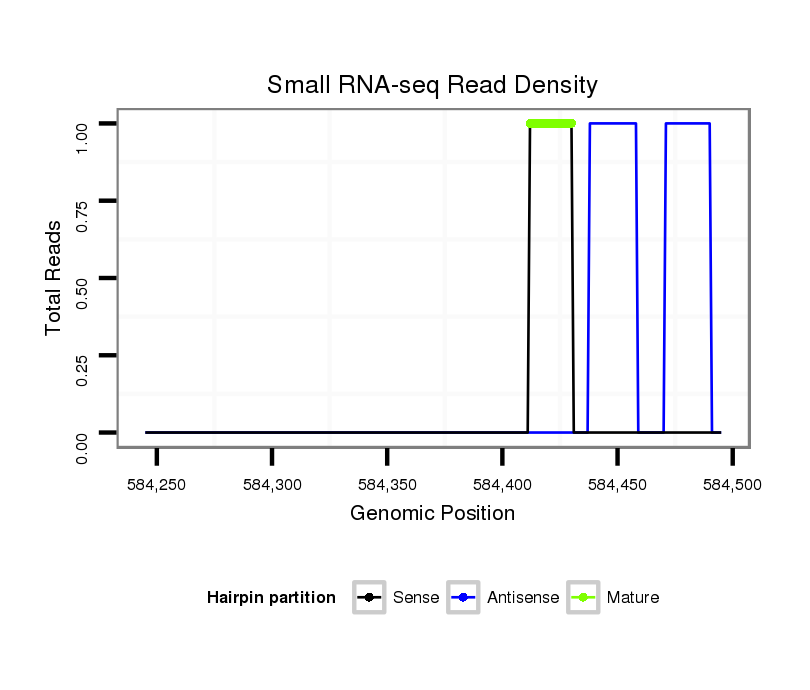
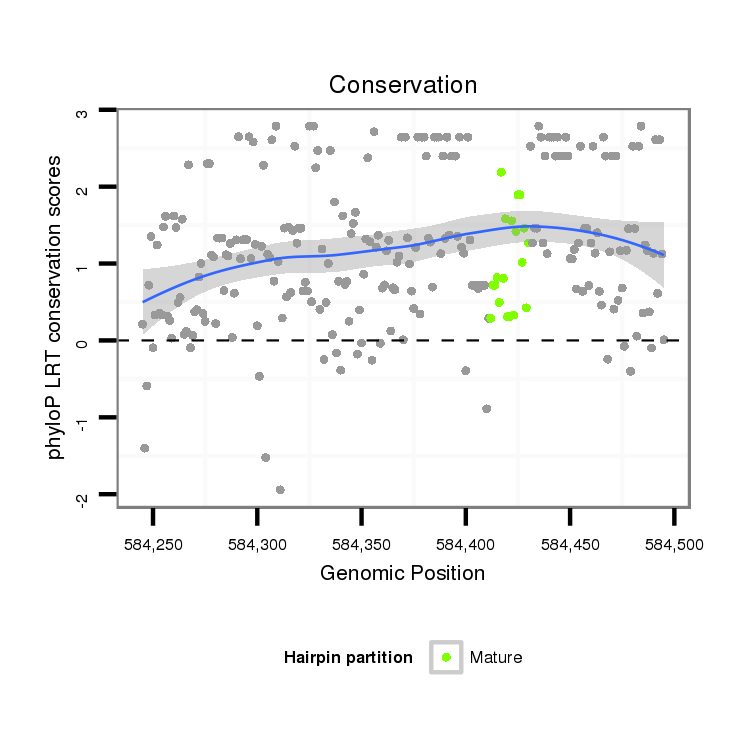
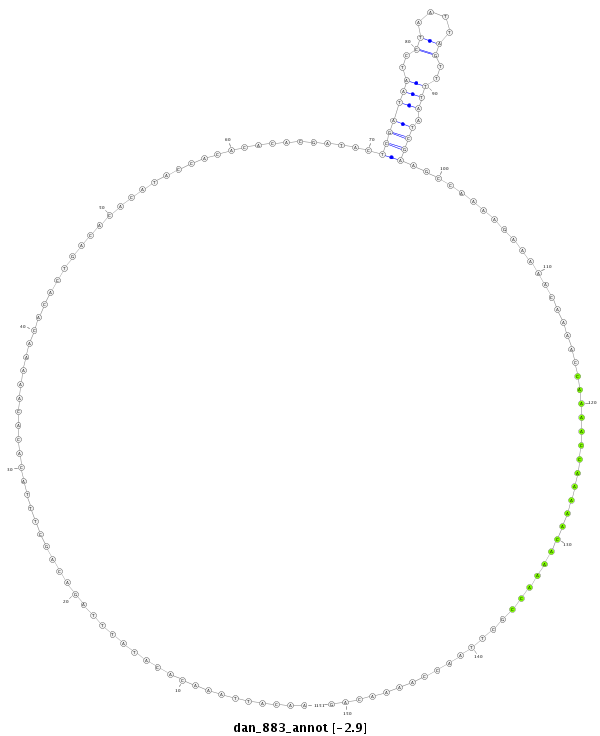
ID:dan_883 |
Coordinate:scaffold_12943:584295-584445 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -2.9 | -2.5 | -2.5 |
 |
 |
 |
CDS [Dana\GF22712-cds]; exon [dana_GLEANR_681:11]; intron [Dana\GF22712-in]
| Name | Class | Family | Strand |
| (CAAAAA)n | Simple_repeat | Simple_repeat | + |
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## ACGAAATGCCGTCTCTCTTGTGAATCAAGCGCCCCTTAATCCATTTACTAAACATTAAACACATATTTAGACAGCTTTACACACAAAAACACACTGACACACATACCACACACACGATACTCGATAATCCTAATTAGTTTTAATCGAAGCCAAAAGAAAAACAAAACCAAAACCAAAAACAAAACCGCTTAACCAAAACAGGCTCGCCACGCTTTGGCTCCTCACCAGTTGGCAACAACAACCCCGATAGC **************************************************......................................................................(((((((..((....))..)).)))))......................................................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M044 female body |
M058 embryo |
V055 head |
V039 embryo |
V106 head |
|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................................................CGCTGAACCATAGCAGGCT............................................... | 19 | 3 | 6 | 8.17 | 49 | 30 | 1 | 11 | 2 | 5 |
| ..........................................................................................................................................................................................GCTGAACCATAACAGGCTAGC............................................ | 21 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................CGCTTAACCATAGCAGGCTA.............................................. | 20 | 3 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................CTTAACCATAGCAGGCTAGC............................................ | 20 | 3 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................CAAAACCAAAAACAAAACC................................................................. | 19 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................................CGCTGAACCATAACAGGCT............................................... | 19 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................GAAAAACACAACCAAAACCA............................................................................ | 20 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................TGACACACGTCCCACACAC.......................................................................................................................................... | 19 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 |
| .................................................................................................................................................................................................CGAAACAGGCTCGACACGC....................................... | 19 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................CGCTGAACCAAACCAGGCT............................................... | 19 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 |
| .......................................................................................................................................................AAAAGAAAAACAACACCAAAA............................................................................... | 21 | 1 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 |
| .........................................................................................................................................................AACAAAAACAAAACGAATACCAAAAACA...................................................................... | 28 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................AAAACCGCTTAACCAGAA..................................................... | 18 | 1 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................................AAAGAACAACAAAACCCAAACC............................................................................. | 22 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................................AGAAAAACAAATCCAACACC............................................................................. | 20 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................................................AAAACAAAAACCAATAACAAAACC................................................................. | 24 | 2 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................AATTAGTTGTAAGCGAAGC..................................................................................................... | 19 | 2 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................GCTTAACCATAGCAGGCT............................................... | 18 | 2 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................................................................CGAAATAGGCTCGACACGC....................................... | 19 | 3 | 14 | 0.21 | 3 | 1 | 0 | 0 | 0 | 2 |
| ........................................................................................................................................................AACGAAAAACACACCCAAAACCA............................................................................ | 23 | 3 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................................ACAAAACCAAAACCCAAAAC....................................................................... | 20 | 1 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................................................................ACGAAACAGGCTCGACACGC....................................... | 20 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................AACCAAAACCAAAAACACCACC................................................................. | 22 | 2 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...............................................CTAAACATTACACCCATA.......................................................................................................................................................................................... | 18 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 |
| .........................................................................................................................................................................................CGCTGAACCATACCAGGCT............................................... | 19 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................AGAAAAACACACCCAAAACCA............................................................................ | 21 | 2 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................................CAAAACCCAAAACAAAACC................................................................. | 19 | 1 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................................AGGCCAAAAGAAAAACAAA...................................................................................... | 19 | 1 | 8 | 0.13 | 1 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................................................................................AAAGAAAAAGAGAACCAACACCA............................................................................ | 23 | 3 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................AACCAAACCCAAAAACAAAA................................................................... | 20 | 1 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................................AAAGAAAAACAACACCAAAACA............................................................................. | 22 | 2 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................AAAAGAAAAACAAAACCGAAA............................................................................... | 21 | 1 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................AAAAAAAACAAAACCAAAACCA............................................................................ | 22 | 1 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................................AAAAAAACAAAACCAAAACC............................................................................. | 20 | 1 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 |
| ...............................................................................................................................................................AAAAAAACCCAAACCCAAAACAAAAC.................................................................. | 26 | 3 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................GAAGCCAAAAGAGAAACAAAATA................................................................................... | 23 | 3 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................AAAAGAAAAACACAACCACCACC............................................................................. | 23 | 3 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................GAAGCCAAAAGAAAAAAAA....................................................................................... | 19 | 1 | 12 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................................................................................AAAGAAAAACAAAGCCGAAAC.............................................................................. | 21 | 2 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................AAAACACAACCAAAAGCAAAAA........................................................................ | 22 | 2 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................AAAACAAGACCAAAACCA............................................................................ | 18 | 1 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................AAAAACACACCCAAAACCAAA.......................................................................... | 21 | 2 | 14 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................AAAAACAAAACCAAAGCCAA........................................................................... | 20 | 1 | 15 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................................ACAAAACGAAAACCAACAACA...................................................................... | 21 | 2 | 15 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................AAAACAACACCCAAACCAAAA......................................................................... | 21 | 2 | 15 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................................AAAAACAAACCCCCCTAACCA........................................................ | 21 | 3 | 15 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................GAAGCCAAAAGAAACACCA....................................................................................... | 19 | 2 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................GAAAAACAGAACCAAAACGA............................................................................ | 20 | 2 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................................CAAAACCAAAAAGAGAACC................................................................. | 19 | 2 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................................ACAAAACCAAAACCAAGAAAAAAA................................................................... | 24 | 2 | 18 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................................AAAGAAAAAGCAAACCCAAACCA............................................................................ | 23 | 3 | 19 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................................AACAAAACCAAAAACAAAAACACCAC.................................................................. | 26 | 3 | 19 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................GAAAAACGAAACCAACACC............................................................................. | 19 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................AATTGAAGCAAAAAGAAAAACC........................................................................................ | 22 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................................................AAAATCAAAACCAAAAACAA..................................................................... | 20 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................................................TTAACCATAGCAGGCTAGC............................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................AAACAACACCCAAACCAAAA......................................................................... | 20 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................GAAAAACAAAACGAACACC............................................................................. | 19 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................GAAAAACAAAACCAAAAACA............................................................................ | 20 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................................AGAAAAACACAACCAAAGC.............................................................................. | 19 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................GAGAAACAAAACCAAACCCCAAA......................................................................... | 23 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
|
TGCTTTACGGCAGAGAGAACACTTAGTTCGCGGGGAATTAGGTAAATGATTTGTAATTTGTGTATAAATCTGTCGAAATGTGTGTTTTTGTGTGACTGTGTGTATGGTGTGTGTGCTATGAGCTATTAGGATTAATCAAAATTAGCTTCGGTTTTCTTTTTGTTTTGGTTTTGGTTTTTGTTTTGGCGAATTGGTTTTGTCCGAGCGGTGCGAAACCGAGGAGTGGTCAACCGTTGTTGTTGGGGCTATCG
**************************************************......................................................................(((((((..((....))..)).)))))......................................................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V105 male body |
M044 female body |
V106 head |
V055 head |
M058 embryo |
V039 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................................................................CGAGGGGTGCGAAAGCGCGGAGT........................... | 23 | 3 | 3 | 402.33 | 1207 | 661 | 387 | 124 | 35 | 0 | 0 |
| .........................................................................................................................................................................................................CGAGGGGTGCGAAAGCGCGGAG............................ | 22 | 3 | 3 | 206.00 | 618 | 390 | 146 | 71 | 11 | 0 | 0 |
| ..........................................................................................................................................................................................................GAGGGGTGCGAAAGCGCGGAGT........................... | 22 | 3 | 3 | 42.33 | 127 | 61 | 48 | 7 | 11 | 0 | 0 |
| ..........................................................................................................................................................................................................GAGGGGTGCGAAAGCGCGGAG............................ | 21 | 3 | 3 | 32.00 | 96 | 50 | 34 | 7 | 5 | 0 | 0 |
| ...........................................................................................................................................................................................................AGGGGTGCGAAAGCGCGGAGT........................... | 21 | 3 | 5 | 16.20 | 81 | 43 | 21 | 11 | 6 | 0 | 0 |
| .........................................................................................................................................................................................................CGAGGGGTGCGAAAGCGCGGA............................. | 21 | 3 | 8 | 13.38 | 107 | 52 | 32 | 16 | 7 | 0 | 0 |
| ...........................................................................................................................................................................................................AGGGGTGCGAAAGCGCGGAG............................ | 20 | 3 | 12 | 11.42 | 137 | 89 | 27 | 14 | 7 | 0 | 0 |
| ..............................................................................................................................................................................................................GGTGCGAAAGCGCGGAGTCG......................... | 20 | 3 | 4 | 11.00 | 44 | 12 | 6 | 17 | 9 | 0 | 0 |
| ............................................................................................................................................................................................................GGGGTGCGAAAGCGCGGAGT........................... | 20 | 3 | 8 | 8.38 | 67 | 33 | 21 | 12 | 1 | 0 | 0 |
| .........................................................................................................................................................................................................CGAGTGGTGCGAAAGCGCGGAGT........................... | 23 | 3 | 1 | 5.00 | 5 | 3 | 0 | 2 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................CGAGGGGTGCGAAAGCGTGGAG............................ | 22 | 3 | 1 | 5.00 | 5 | 1 | 0 | 0 | 0 | 4 | 0 |
| .........................................................................................................................................................................................................CGAGGGGTGCGAAATCGCGGAGT........................... | 23 | 3 | 1 | 4.00 | 4 | 1 | 3 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................CGAGGGGTGCGAAAACGCGGAGT........................... | 23 | 3 | 2 | 4.00 | 8 | 0 | 7 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................CGAGGGGTGCGAAAGCGAGGAG............................ | 22 | 2 | 1 | 3.00 | 3 | 2 | 0 | 0 | 1 | 0 | 0 |
| .........................................................................................................................................................................................................CGAGGGGTGCGAAAGCGAGGAGT........................... | 23 | 2 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................GGGTGCGAAAGCGAGGAGTCG......................... | 21 | 3 | 1 | 3.00 | 3 | 2 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................AGCGGTGCGAAAGCGCGGAGTCG......................... | 23 | 3 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................CGAGCGGTGCGAAAGCGCGGAGTCG......................... | 25 | 3 | 1 | 2.00 | 2 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................CGAGGGGTGCGAAACCGCGGAGTC.......................... | 24 | 3 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................CGAGGGGTGCGAAACCGCGGAGT........................... | 23 | 2 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................CGGGGTGCGAAACCGAGGAGTCG......................... | 23 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .........................................................................................................................................................................................................CGAGGGGTGCGAAAGCGTGGA............................. | 21 | 3 | 2 | 1.00 | 2 | 0 | 0 | 1 | 0 | 1 | 0 |
| ..........................................................................................................................................................................................................GAGCGGTGCGAAAGCGCGGAGTCG......................... | 24 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................TCAACCGTTGTTGTTGGGGC..... | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................CGAGGGGTGCGAAAGCGTGGAGT........................... | 23 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................GCGGTGCGAAAGCGTGGAGTCG......................... | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................GAGGGGTGCGAAAGCGAGGAGTCG......................... | 24 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................CGAGCGGTGCGAAAGCGCGGAGT........................... | 23 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................CGAGTGGTGCGAAAGCGCGGAG............................ | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................ACGAGGGGTGCGAAACCGCGGAG............................ | 23 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................ATTGGTTTTGTCCGCGCGATG......................................... | 21 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................GGGGTGCGAAAGCGAGGAGTC.......................... | 21 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................ACGAGGGGTGCGAAACCGCGGAGT........................... | 24 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................GTGCGAAACCGTGGAGTGG......................... | 19 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................................................................................................................................GGTGCGAAAGCGAGGAGTCG......................... | 20 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................CGAGGGGTGCGAAAGCGCGGAGTGG......................... | 25 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................GCGGTGCGAAAGCGCGGAGTC.......................... | 21 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................CGAGGGGTGCGAAAGCGAGGAGTCG......................... | 25 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................ATGGCGTGTGTGCAATAAGCT............................................................................................................................... | 21 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .............................................................................................................................................................................................................CGGTGCGAAAGCGCGGAGTCG......................... | 21 | 3 | 2 | 1.00 | 2 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................GAGGGGTGCGAAATCGCGGAGT........................... | 22 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................GAGGGGTGCGAAACCGCGGAGT........................... | 22 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................GTTTTGTCCGAGCGGTGCGAA..................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................GCGGTGCGAAAGCGCGGAGTCG......................... | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................AATTGGTTCTGTCCGGGCGG........................................... | 20 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| .........................................................................................................................................................................................................CGAGGGGTGCGAAAGCGCGGAGTG.......................... | 24 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................GAGGGGTGCGAAAGCGCGGA............................. | 20 | 3 | 17 | 0.53 | 9 | 5 | 2 | 2 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................GAGGGGTGCGAAAACGCGGAGT........................... | 22 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................ATGGGTTTTGTCCGCGCGATG......................................... | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................GGGGTGCGAAATCGCGGAGT........................... | 20 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................GGGGTGCGAAAGCGCGGAGTG.......................... | 21 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................CCGAGGGGTGCGAAAGCGCGGAGT........................... | 24 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................CGAGGGGTGCGAAAACGCGGAG............................ | 22 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................GGTGCGAAAGCGTGGAGTC.......................... | 19 | 3 | 9 | 0.33 | 3 | 1 | 0 | 0 | 0 | 2 | 0 |
| ..............................................................................................................................................................................................................GGTGCGAAAGCGCGGAGTC.......................... | 19 | 3 | 14 | 0.29 | 4 | 0 | 4 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................GGTGCGAAAGCGCGGAGT........................... | 18 | 2 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................CGGTGCGAAAGCGTGGAGTC.......................... | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..........................................................................................................................................................................................................GAGGGGTGCGAAATCGCGGA............................. | 20 | 3 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................GAGGGGTGCGAAAGCGGGGAG............................ | 21 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................ACATTGGTTTTGTCCGCGCG............................................ | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................CGAGTGGTGCGAAAGCGCGG.............................. | 20 | 3 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................CCGTTGTTGGTGGTGCTA... | 18 | 2 | 11 | 0.09 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .........................................................................................................................................................................................................CGAGGGGTGCGAAAGCGTGG.............................. | 20 | 3 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................ATGAGTGGTCAAGCGTTGA.............. | 19 | 3 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................AGAGGGGTGCGAAAGCGAGG.............................. | 20 | 3 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................GAGTCGTCGACCATTGTTG............ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .............................................TGGATTTGTAGTTTGTGTAT.......................................................................................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ........................................................................................................................................................................................................CGGACCGGTGCGAATCCGA................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droAna3 | scaffold_12943:584245-584495 + | dan_883 | ACGA----------------AATGCCGT------CTCTCTTGTGAATC-----AAGCG-CCCC--TTAATCCATTTACTAAACATTAAAC---------ACA--TATTTAGACAGCTTTACACA------------------------------------CAAAAAC--ACAC-------------------------------------TGACACACATACC----------ACACACAC--------GA---TAC-----TCG-------ATA---AT-CCTAA-TTAGTTTT-AATCGA--AGCCAAA-A--------------GAAAAACAAAACCAAAACCAAAAACAAAACCGCTTAACCAAAACAGGCTCGCCACGCTTTGGCTCCTCACCAGTTGG---------CA----------------------------------------------------------ACAACAAC--------------------------------------C---------------CCGATAGC |
| droBip1 | scf7180000396535:144748-144996 - | ----------------------------TGACAACTCTCTCGTTAAGC-----GAGTGCCCCC--TTAATCCATTTACTAAA----------------------TACATAGACAGCTTTACACA------------------------------------CACAAAACCACAC-------------------------------------TGACACACATGCC----------ACACACAC----ACACGA---TAC-----TCGATATCCTATA---AT-CTTAA-TTAGTTTTAAATCGA--AGCCAAA-A--------------GAGAAACAAATCCAAAACCAAAACCAAAACCGCTTAACCAAAACAGGCTCGCCACGCTTTGGCTCCTCACCAGTTGG---------CA----------------------------------------------------------ACAACAAT--------------------------------------C---------------CCGATAGC | |
| droKik1 | scf7180000302679:1114691-1114936 + | ATGG-------GA---------------------ATCTCATGGGATTCAACACAAGCG-CCCC--TTAATCCACATACTAAACATTAAAA--------AACA--TA-AGAGACAGCTTAACACA------------------------------------CACACAC--ACACA------------CAC-------A--------------------CACACCA--CA--AACACACACAC----AAA------CAG-----TTG-------ATG---AT-CTGAACCTAGTTTT-AATCAAA-AGCCAA--C--------------AAA-------------AC---------AACCGCTTAACCAAAACAGGCTCGCCACGCTTTGGCTCCTCGCCGGGCAT---------CA----------------------------------------------------------GCAGCAGCAGCAACA---ACCAC-----------------------A---------------CCGATAGC | |
| droFic1 | scf7180000453712:31616-31881 + | ATGA-------AT--A--CAATTGTAGT------------TGTGATTT-----AAGCA-CCCC--TTAATCCACATACTAAACATCAAAC--------AACA--TACCTAGACAGCTTTAAACA------------------------------------CACACAC--A--AA---------ACACACACCACACACACACACTACTCA-----CACATACC----------ACATACAA----AAA-GAGACCAC-----CTG-------ATA---AT-CTGAA-CTAGTTTT-AATCAA--AGCCAAA-C--------------GAA-------------ACCA--ACCAAAACCGCTTAACCAAAACAGGCTCGCCACGCTTTGGCTCCTCGCCGGTCAG---------CA----------------------------------------------------------GCAACAAC------------AAC-----------------------A---------------CAGATAGC | |
| droEle1 | scf7180000491186:2178052-2178322 + | A--------------------------------TACTTCTTGTGATTT-----AAGCA-CCCC--TTAATCCACATACTAAACATTCAAT--------TACA--TACTTAGGCAGCTTTACACA------------------------------------CACAAAC--ACATATAC------------TTTACACACACACTTTACACA-----CACACACT----------CCATACAA------A------CAC-----TTG-------ATA---AT-CTGAA-CTAGTTTT-AATCAA--AGCCAAAACGAAAAAAATTAAAAAAAAAACAAA---AAAACCA--ACCAAAACCGCTTAACCAAAACAGGCTCGCCACGCTTTGGCTCCTCAACGGGCAG---------CA----------------------------------------------------------GCAACAAC--------------------------------------A---------------CTGATAGC | |
| droRho1 | scf7180000778060:38436-38685 - | ACAAAGTATGGTA-TT--CAAATGTAGTTGACATATCTCTTGTGATTT-----AAGCA-CCCC--TTAATCCACATACTAAACATTAAAA--------TACA--TACTTAGACAGCTTTACACA------------------------------------CACACAC--ACA-------------------------------------------CACATATC----------ACATACAA------A------CAC-----TTG-------ATA---AT-CTGAA-CTAGTTTT-AATCAA--AGCCAAA-C--------------GAA------------AACCA--ACCAAAACCGCTTAACCAAAACAGGCTCGCCACGCTTTGGCTCCTCACCGGGCAG---------CA----------------------------------------------------------GCAACAAC--------------------------------------A---------------CTGATAGC | |
| droBia1 | scf7180000302408:3972199-3972427 - | GCGA-------------------------GACATATCTCTTGTGACCT-----AAGCA-CCCC--CTAATCCACATACTAAACATTCAAG--------AACA--TACCTAGATAGCTTTACACACACTCTCACA--------------------------------------CA---------ACACAT-------------------------------AC--------------CACAC------A------CAC-----TTG-------ATGATAAT-CTGAA-CTAGTTTT-AATCAA--AGCCAAA-C--------------GAA-------------ACCA--ACCAAAACCGCTTAACCAAAACAGGCTCGCCACGCTTTGGCTCCTCACCGGGCGG---------CA----------------------------------------------------------GCAACACC--------------------------------------A---------------CTGATAGC | |
| droTak1 | scf7180000415702:273927-274168 - | ATT---------------CAAGTGTAGTTGACATATCTCTTGTGATTT-----AAGCA-CCCCTCTTAATCCACATACTAAACATTAAAC--------TACA--TACTTAGACAGCTTTACACA------------------------------------CACACAC--A--TA---------A--------------------------------------------------------C------A------CAC-----TTG-------ATA---AT-CTGAA-CTAGTTTT-AATCAAAAAGCCAAA-C--------------GAA-------------ACCA--ACCAAAACCGCTTAACCAAAACAGGCTCGCCACGCTTTGGCTCCTCACCGGGCAG---------CA----------------------------------------------------------ACAGCAGCAGCAACAACAACACC-----------------------A---------------CTGATAGC | |
| droEug1 | scf7180000409554:224582-224851 - | GT----------------CGAATGTAGTTGACATATCTCTTGTGATTT-----AAGCA-CCCC--TTAATCCATATACTAAACATTCAAA--------TACA--TACCTAGACAGCTTTACACA------------------------------------CACACAC--AC-------------CACAC-------AC------------TCACAAACATAAC----------ACATACAA----ACA------CAC-----TTG-------ATA---AT-CTGAA-CTAGTTTT-AATCAA--AGCCGAAACAAAA----C-----AAA-------------ACCA--ACCAAAACCGCTTAACCAAAACAGGCTCGCCACGCTTTGGCTCCTCACCGGGCAG---------CA----------------------------------------------------------GCAACAACAACAACAACAACAAC-----------------------A---------------CTGATAGC | |
| dm3 | chr2L:9244069-9244312 + | ATGA-------AA-AA--CAAATGTAGTTGATATAAATCTTGTGATTT-----AAGCA-CCCC--TTAATCCACATACTAAACATTAAAC--------TACA--TACTTAGACAGCTTTACACACACTCACACA--------------------------------------TA---------ACATAC-------A--------------------CA---CA--CC----CACACACAC------A------CAC-----TTG-------ATG---AC-CTGAA-CTAGTTTT-AATCAA--TGCCAAA-C--------------GAA-------------ACCA--------ACCGCTTAACCAAAACAGGCTCGCCACGCTTTGGCTCCTCACCGGGCAG---------CA----------------------------------------------------------GCAACAAC--------------------------------------A---------------CTGATAGT | |
| droSim2 | 2l:8936812-8937048 + | dsi_2498 | AC-----------------AAATGTAGTTGATATATCTCTTGTGATTT-----AAGCA-CCCC--TTAATCCACATACTAAACATTAAAC--------TACA--TACTTAGACAGCTTTACACACACTCACACA--------------------------------------TA---------ACACAC------------------------------------A------CACACACAC------A------CAC-----TTG-------ATG---AC-CTGAA-CTAGTTTT-AATCAA--AGCCAAA-C--------------GAA-------------ACCA--ACCAAAACCGCTTAACCAAAACAGGCTCGCCACGCTTTGGCTCCTCACCGGGCAG---------CA----------------------------------------------------------GCAACAAC--------------------------------------A---------------CTGATAGT |
| droSec2 | scaffold_3:4703050-4703295 + | ATGA-------AG-AA--CAAATGTAGTTGATATATCTCTTGTGATTT-----AAGCA-CCCC--TTAATCCACATACTAAACATTAAAC--------TACA--TACTTAGACAGCTTTATACACACTCACACA--------------------------------------TA---------ACACAC-------A---------------------------CA------CACACACAC------A------CAC-----TTG-------ATA---AC-CTGAA-CTAGTTTT-AATCAA--AGCCAAA-C--------------GAA-------------ACCA--ACCAAAACCGCTTAACCAAAACAGGCTCGCCACGCTTTGGCTCCTCACCGGGCAG---------CA----------------------------------------------------------GCAGCAAC--------------------------------------A---------------CTGATAGT | |
| droYak3 | 2L:11911309-11911564 + | ACAAGTACG-ATA-AA--CAAATGTAGTTGATATATCTCTTGTGATTT-----AAGCA-CCCC--TTAATCCACATACTAAACATTAAAC--------TACA--TACCTAGACAGCTTTACACACACTCACACA--------------------------------------TA---------ACACAC-------A-----------------------ACCA--CA----CACACACAC------A------CAG-----TTG-------ATG---AC-CTGAA-CTAGTTTT-AATCAA--AGCCAAA-C--------------GAA-------------TCCA--ACCAAAACCGCTTAACCAAAACAGGCTCGCCACGCTTTGGCTCCTCACCGGGCAG---------CA----------------------------------------------------------GCAACAAC--------------------------------------A---------------CTGATAGT | |
| droEre2 | scaffold_4929:9849629-9849868 + | AA----------------CAAACGTAGTTGATATATCTCTTGCGATTT-----AAGCA-CCC----TAATCCACATACTAAACATTAAAC--------TACA--TACTTAGACAGCTTTACACAAACTCACACA--------------------------------------TA---------GTACAC-------A---------------------------CA------CACAAACAC------A------CAC---TTTTG-------ATG---AC-CTGAA-CTAGTTCT-AATCAA--AGCCAAA-C--------------GAA-------------ACCA--ACCAAAACCGCTTAACCAAAACAGGCTCGCCACGCTTTGGCTCCTCACCGGGCAG---------CA----------------------------------------------------------GCAACAAC--------------------------------------A---------------CTGATAGC | |
| dp5 | 4_group5:1563288-1563548 + | ACTAGCTCGG-GAGTTCC------CCGTTTATAGA-------TCAGTT-----AGGCG-CCCC--TTAATCCATATACTAAACATCCA------------CATTTACTTAGACAGCTTCACACA------------------------------------CACAACC--AGACACACACACAACCAGAC-------AC--------------A-----------CACACAAACACACATAC------A------CAC-----TT---------TA---AT-CTCGA-TTAGTTTT-AATCAA--AGCCAAA-C--------------CAA-------------ACC---------ACCGCTTAACCAAAACAGGCTCGCCACGCTTCGGCTCCTCCCCGGGCAC---------GG----------------------------------------------------------CCGCCAACAGCAGCA---GCCAC-----------------------AGTCACAGC--------------C | |
| droPer2 | scaffold_5:5971398-5971642 + | ACTAGCTCGG-GAGTTCT------CCGTTTATAGA-------TCAGTT-----AGGCG-CCCC--TTAATCCATATACTAAACATCCA------------CATTTACTTAGACAGCTTCACACA------------------------------------CACAACC--AGACA------------CAC-------A------------------CACA-----------AACACACATAC------A------CAC-----TT---------TA---AT-CTCGA-TTAGTTTT-AATCAA--AGCCAAC-C--------------CAA-------------ACC---------ACCGCTTAACCAAAACAGGCTCGCCCCGCTTCGGCTCCTCCCCGGGCAC---------GG----------------------------------------------------------CCGCCAACAGCAGCA---GCCAC-----------------------AGTCACAGC--------------C | |
| droWil2 | scf2_1100000004521:249658-249943 + | dwi_1196 | A------------------------------------------------------GCT-CCCCACACTGCCCATATACTAAACATTAAATACACAAAAAACA--CACTTAGACAGCTTCGCACA------------------------------------CACACAC--ATACATACAGAAAGACATAC-------AA--------------A---------CA--CA----CACATACAC------A------CAC-----AAA--ATACCTTA---AT-TATGA-TTAGCGAT-AACCAAA-AGCCAAAACCAAAAAAACAAAAAAAA----------------------AAAAACGCTTAACCGAAACAGGCTCGCCACGCTTTGGTGCATCTCCTGGCACGAATACCGGCAACAACAACAACAGC-A------------------------------------------GCAACAAC------------AAT-----------------------AGCAACAGCAACCATACTGATAAT |
| droVir3 | scaffold_12963:650912-651175 + | dvi_9891 | -------------------------------------------------------------CC--CTAATTACTGTACTAAATATTAA----------------TACATAAACAGCTTCACACACACACACACACACGAGCATACCAACACCGTTTCAAACACACAC--A--------------CACAC-------A-------------------------CACA------CACACACACACATACA------CGCAGACATTG-------TTA---ATATTTGA-CTAGTTTTTAACCAA--AGCCAAA-C--------------AAA-------------TCA---------AACGCTTAACCGAAACAGGCTCAACGCGTTTCGGTTTGTCGCCTGGCAA---------CA----------------------------------------------------------ACAACAACAACAACAGCGGCAAC-----------------------AACCACAACAATCACAATGATAGT |
| droMoj3 | scaffold_6540:26135234-26135480 - | CCGA-------CT---------------------GT------CCAACT-----GGTGT-CCAA--CTAACCCACACAGCTGACACAGCAC--------AGAA--AA--CGAAAAGCACCACAGA------------------------------------CAAGGCC--A--AA---------A------------------------------------------------CACCCACCA------A----------------------------------------------------------------------------------------------------------TGCTGTCAA-T--------------GGTGCCACAGTTC---------------CATCCGCAACACCAACAACAACCAGTAATAATAACAACACGAACTCGAACTCATCCACTACTACTACCAACAACAACAACACCATCCCCTCTGCTCTCTCTAACCTCTCTCT--GCAC-GACACGGCAGCGACAGC | |
| droGri2 | scaffold_9046:1006-1171 - | AC-------------------------------------------------------------------------------------------------ACA--TACCAACACCGTTTCGCAAA------------------------------------CACACAC--ACACA---------CCA--------------------------A---------CACA------CACACACAC------A------CAT-----TTG-------TTA---ATATTTGA-TTAGTTTTTAACCAA--AGCCAAA-C--------------AAA-------------TCA---------AACGCTTAACCGAAACAGGCTCAACGCGTTTCGGTTTGTCGCCTGGCAA---------CA----------------------------------------------------------ACAACAAC--------------------------------------------------------AACAGC |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/17/2015 at 11:29 PM