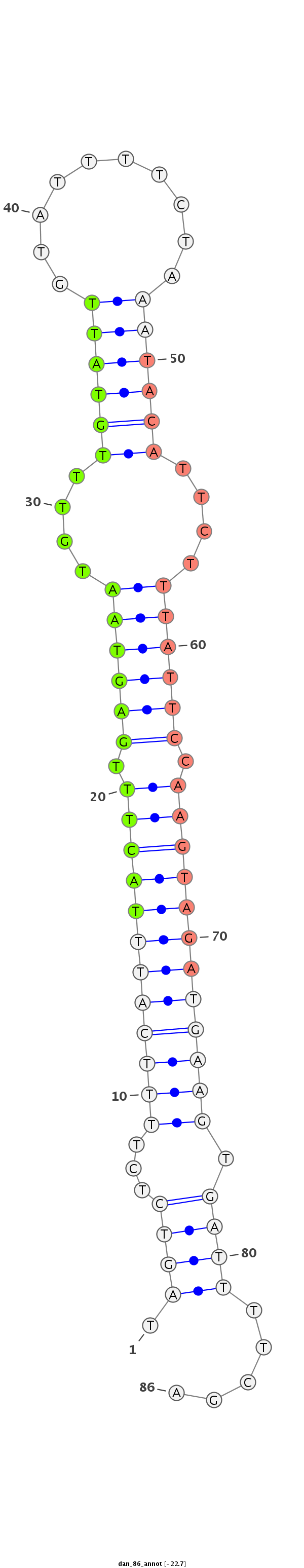
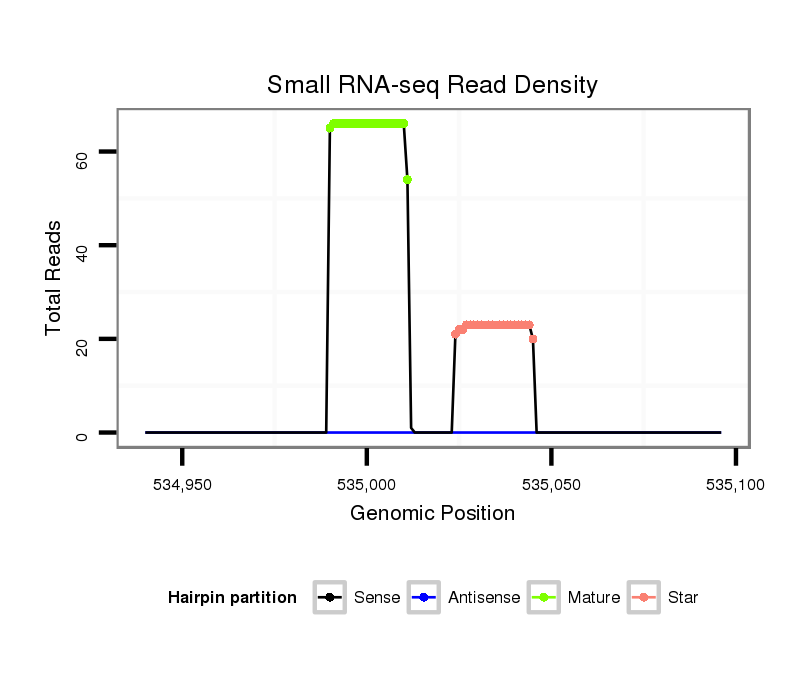
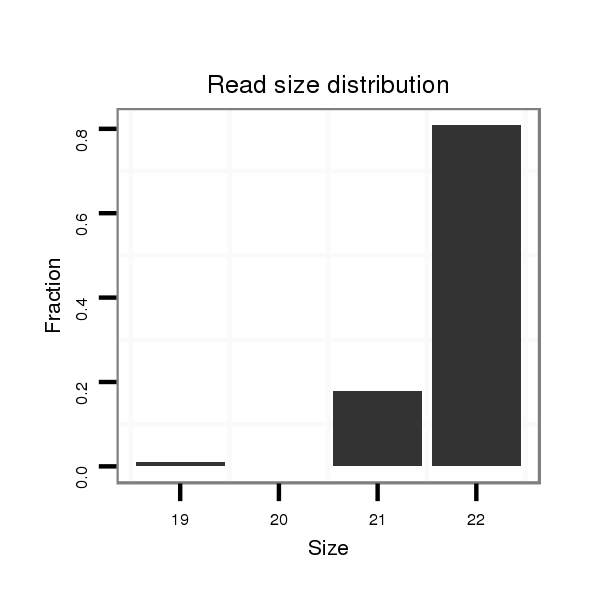
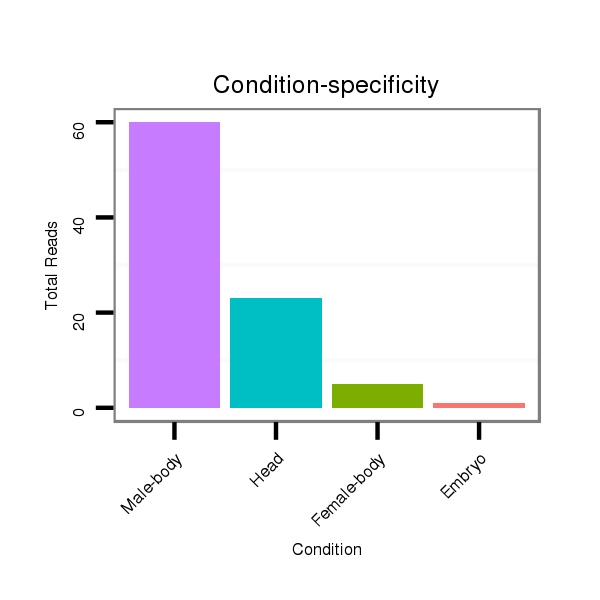
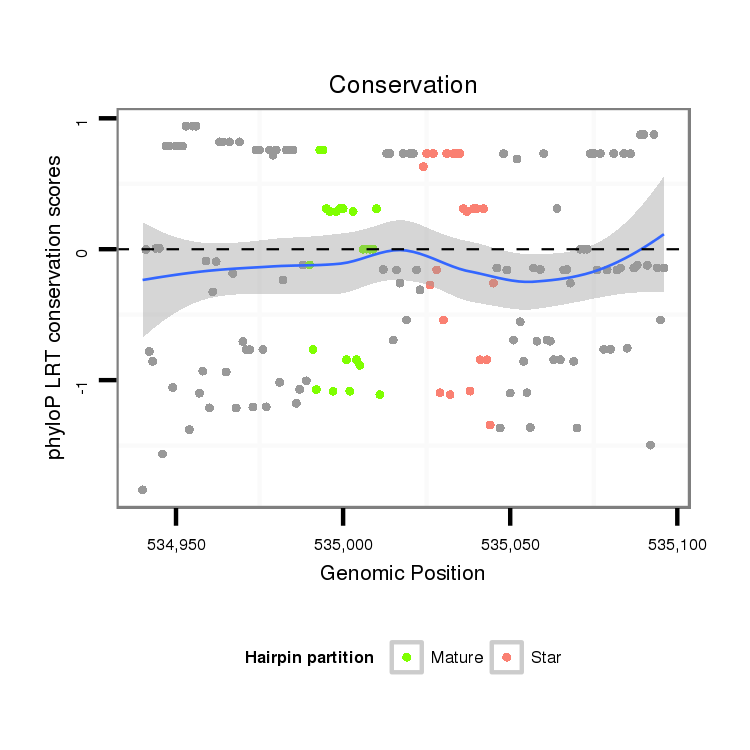
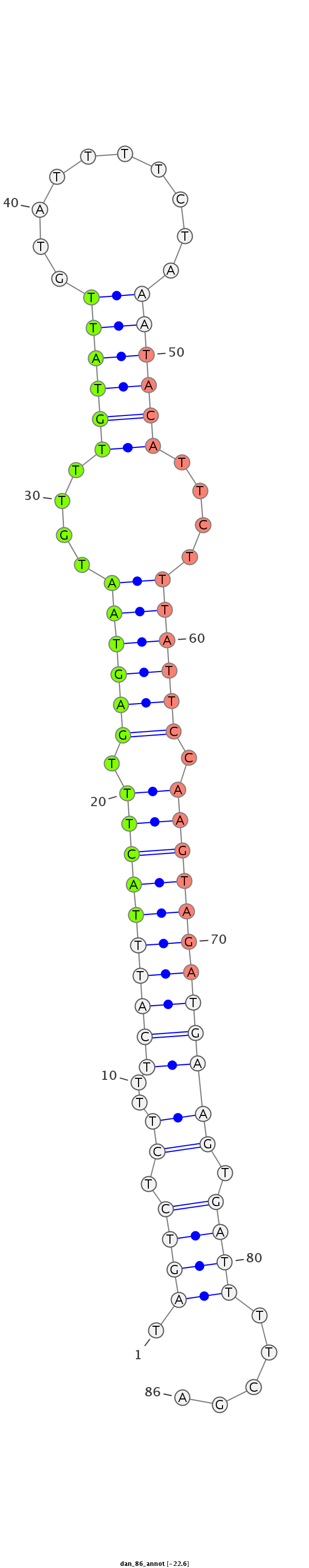
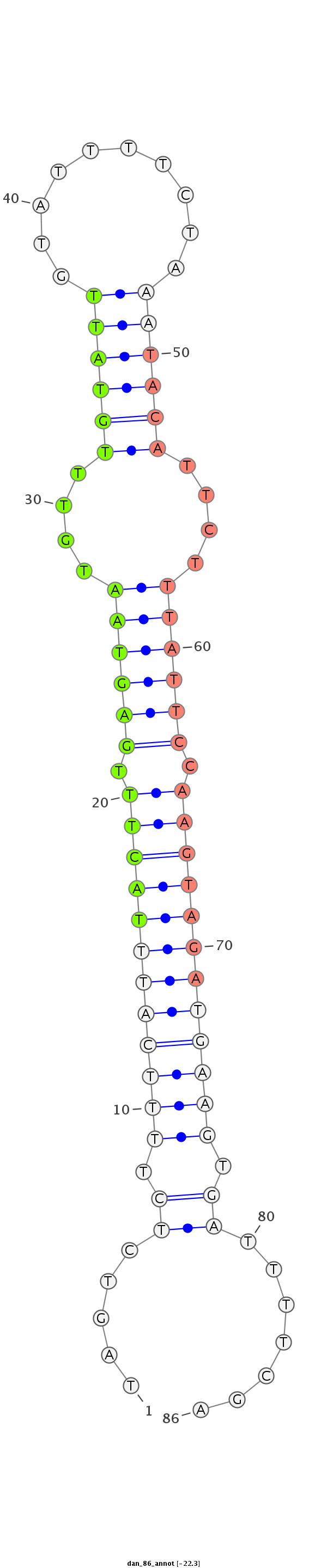
ID:dan_86 |
Coordinate:scaffold_12903:534990-535046 + |
Confidence:confident |
Class:Testes-restricted |
Genomic Locale:intergenic |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -22.6 | -22.3 | -22.3 |
 |
 |
 |
intergenic
No Repeatable elements found
|
AATAAATATCATAAAAATAAATATTAAATATTACTTAGTCTCTTTTCATTTACTTTGAGTAATGTTTGTATTGTATTTTCTAAATACATTCTTTATTCCAAGTAGATGAAGTGATTTTCGATCATCAGTTGAAATAAAAAAAATAAATATAACAACT
***********************************.((((...((((((((((((.((((((....((((((..........))))))....)))))).)))))))))))).)))).....************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | V105 male body |
V106 head |
M044 female body |
V055 head |
M058 embryo |
|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................TACTTTGAGTAATGTTTGTATT..................................................................................... | 22 | 0 | 1 | 53.00 | 53 | 41 | 8 | 3 | 1 | 0 |
| ....................................................................................TACATTCTTTATTCCAAGTAGA................................................... | 22 | 0 | 1 | 18.00 | 18 | 5 | 11 | 2 | 0 | 0 |
| ..................................................TACTTTGAGTAATGTTTGTAT...................................................................................... | 21 | 0 | 1 | 12.00 | 12 | 12 | 0 | 0 | 0 | 0 |
| ..................................................TACTTTGAGTAATGTTTGTATTA.................................................................................... | 23 | 1 | 1 | 4.00 | 4 | 1 | 3 | 0 | 0 | 0 |
| ....................................................................................TACATTCTTTATTCCAAGTAG.................................................... | 21 | 0 | 1 | 3.00 | 3 | 1 | 2 | 0 | 0 | 0 |
| ..................................................TACTTTGAGTAATGTTTGTATTT.................................................................................... | 23 | 1 | 1 | 3.00 | 3 | 2 | 0 | 0 | 1 | 0 |
| ..................................................TACTTTGAGTAATGTTTGTATTAAA.................................................................................. | 25 | 2 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 |
| ................................................TCTACTTTGAGTAATGTTTGTATAA.................................................................................... | 25 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 |
| ..................................................TACTTTGAGTAATGTTTGTATTGA................................................................................... | 24 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................TACTTTGAGTAATGTTTGTAA...................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................TACATTCTTTATTCCAAGTAAA................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................TACTTTGAGTAATGTTTGTAGT..................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................TACATTCTTTATTCGAAGTAGA................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...................................................ACTTTGAGTAATGTTTGTATTG.................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................GACATTCTTTATTCCAAGTAGA................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................................ATTCTTTATTCCAAGTAGA................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 |
| ..................................................TACTTTGAGTAATATTTGTAT...................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................ACATTCTTTATTCCAAGTAGA................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................TTACAGATTAGTTAGTCTCTT................................................................................................................. | 21 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 |
| ....................................................TACTGAGTAATGTTTGTAT...................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
|
TTATTTATAGTATTTTTATTTATAATTTATAATGAATCAGAGAAAAGTAAATGAAACTCATTACAAACATAACATAAAAGATTTATGTAAGAAATAAGGTTCATCTACTTCACTAAAAGCTAGTAGTCAACTTTATTTTTTTTATTTATATTGTTGA
************************************.((((...((((((((((((.((((((....((((((..........))))))....)))))).)))))))))))).)))).....*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M058 embryo |
V039 embryo |
V055 head |
|---|---|---|---|---|---|---|---|---|
| ..........................TTAATATGAATCAGAGAAGA............................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| ..................................................................CCAGAACAGAAAAGATTTAT....................................................................... | 20 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| ..................................................ACGTACCTCATTACAAACA........................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 |
| Species | Coordinate | ID | Type | Alignment |
|---|---|---|---|---|
| droAna3 | scaffold_12903:534940-535096 + | dan_86 | confident | AATA----AATATCATAAAAATAAAT---------ATTAAATA------------------------------------TTACTTAGTCTCTTTTCATTTACTTTGAGTAATGTTTGTATTGTAT---TTTCTAAATAC----ATTCTTTATTCCAAGTAGATGA-AGTGATTTTCGAT----CATCAGTTGAAATA---AAAAAAAT--AAATATAACAACT |
| droBip1 | scf7180000395952:2268-2439 - | CCTA----GGC----------------------------------ATCTAATAAAATCTTTAAAAAATGTGTACAAATACATATTTTTCTATTTTGGTGTTTTTTGCGTAGGGCA----TAGTACTTTTTTTTAA--AC----ATCTTATATTCAAAATGCATTAAAACGGGCGTTGACTCAGTGTTAGTGT---TAAAAAATATAATAAATATATAAAATTT | ||
| droRho1 | scf7180000778091:407267-407291 + | CAGC----AATATAATAACAAT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATAATAT-- | ||
| droTak1 | scf7180000415843:978318-978480 - | TATTTTTTAACATGATAAGAAAGGCTATTTAATTCTTTTACGAATATTTAATAAAAATATTCAATGCTGCATTTAAAC------------------------------------------CATAT---AGTCTATGTAATCCAAAACTCTAT---------CCCAATTTGGGACCTAATTAAAT---TAGGC---TAATTTATTTATCCAAAACGTAGAATCC | ||
| droEre2 | scaffold_4690:9432332-9432380 + | AAAA----AAA------AAAAGTGGC---------TTTGACCA------------------------------------TTATTTAATCTAGTTTCTCTCAATT----------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 10/20/2015 at 11:18 PM