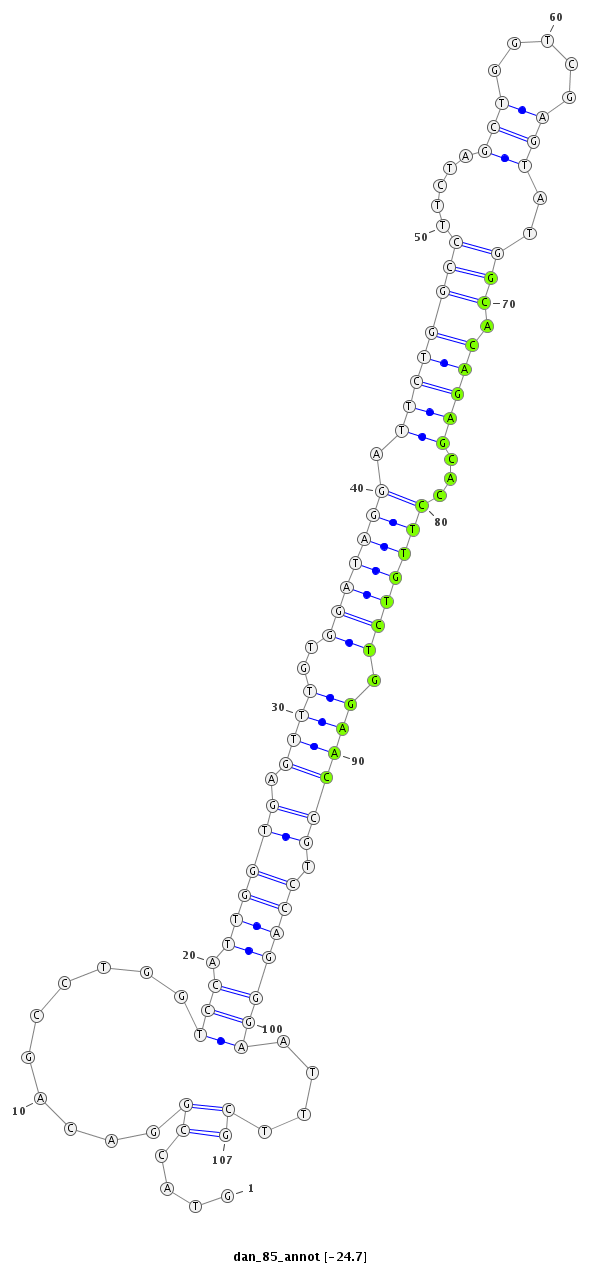
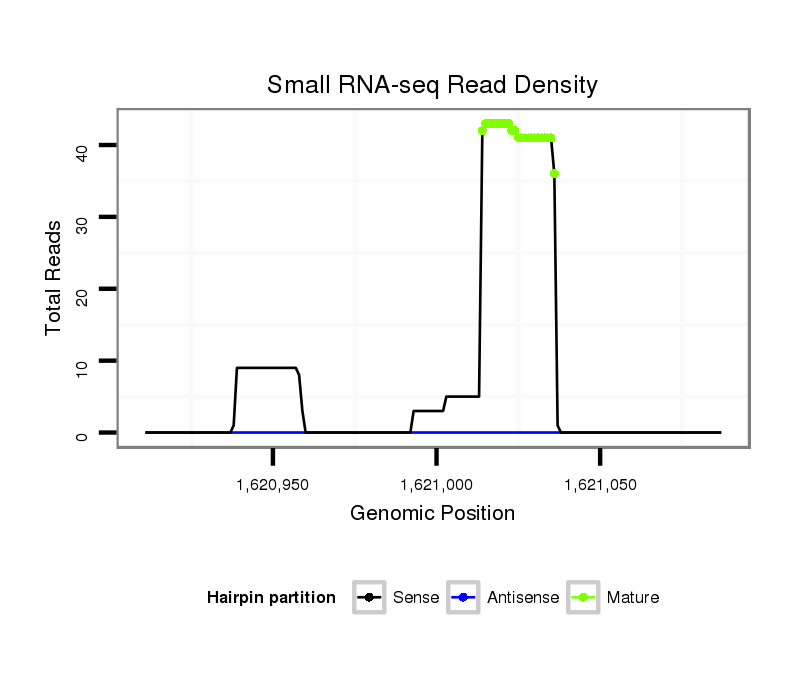
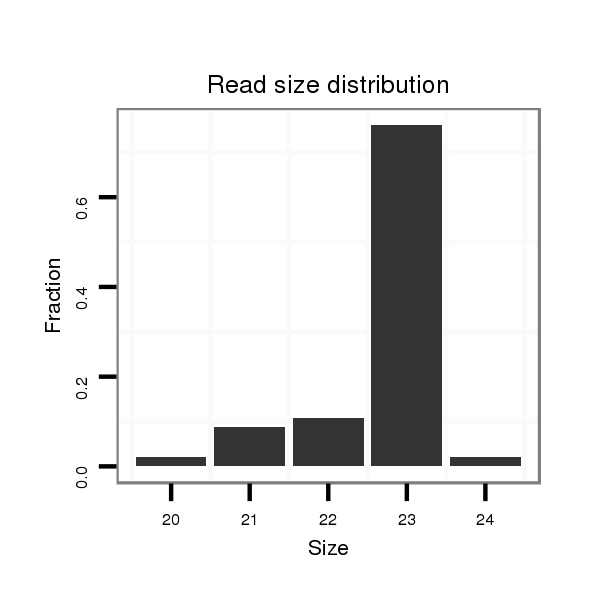
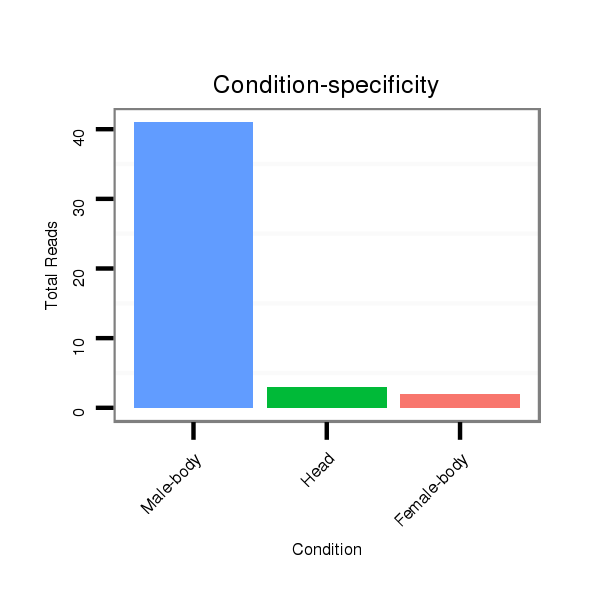
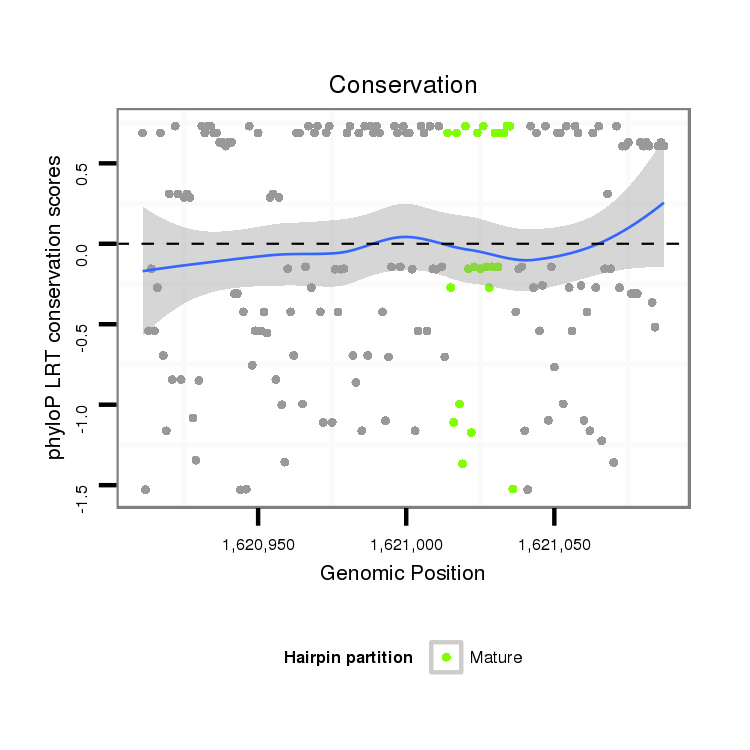
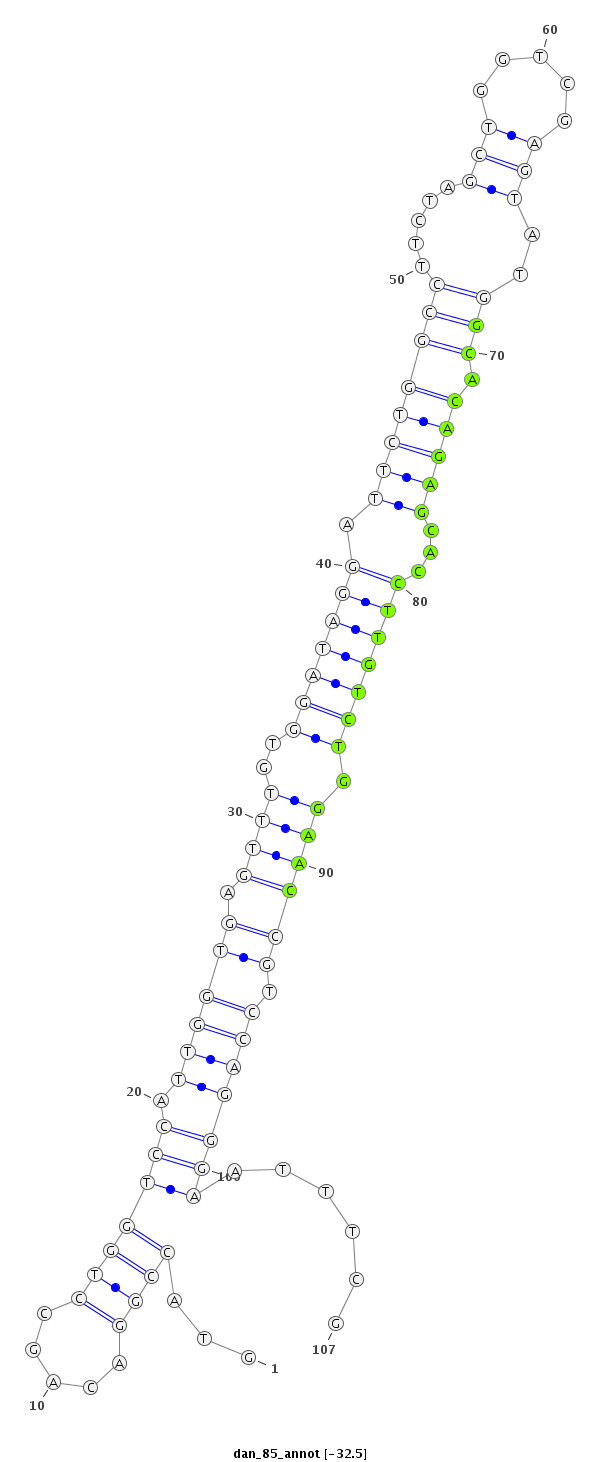
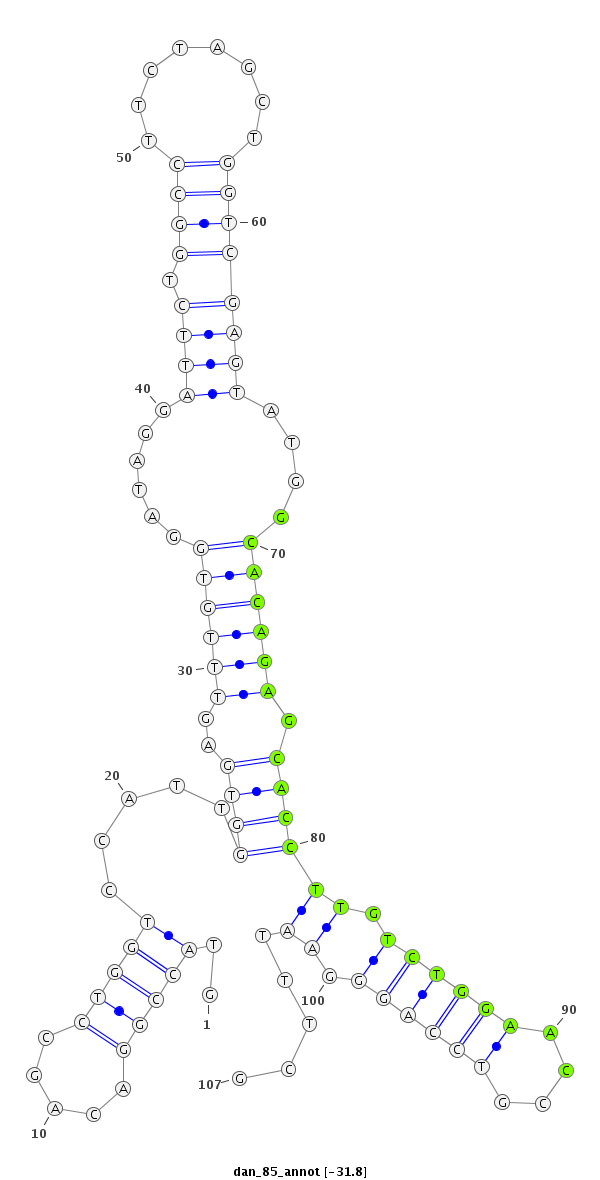
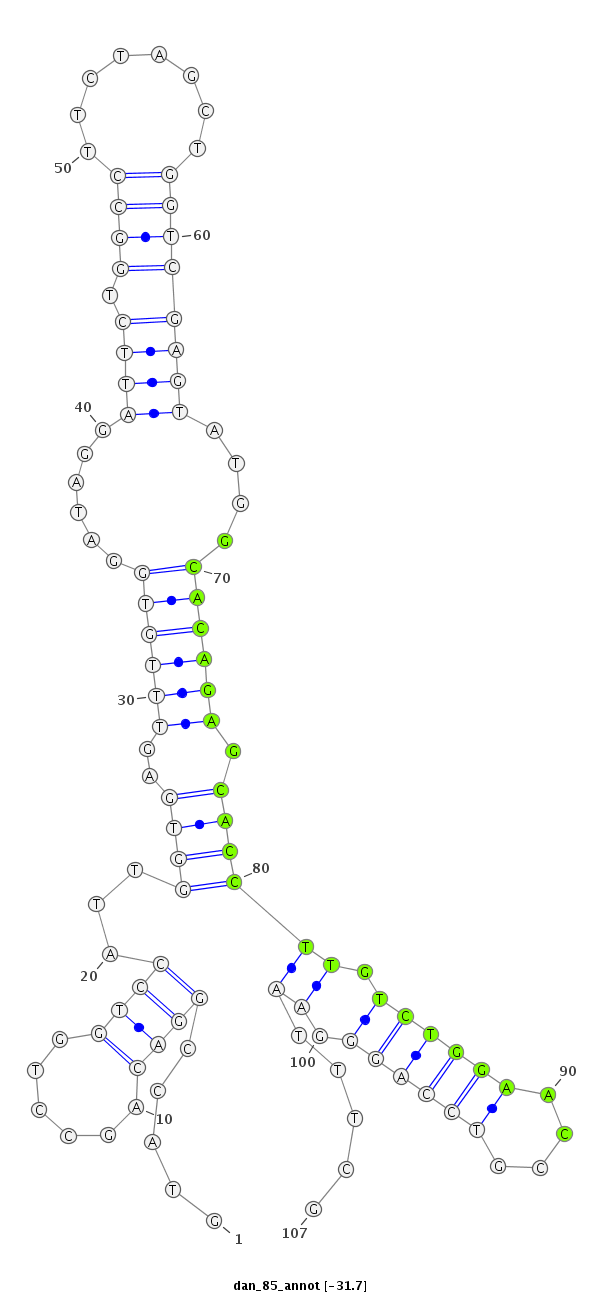
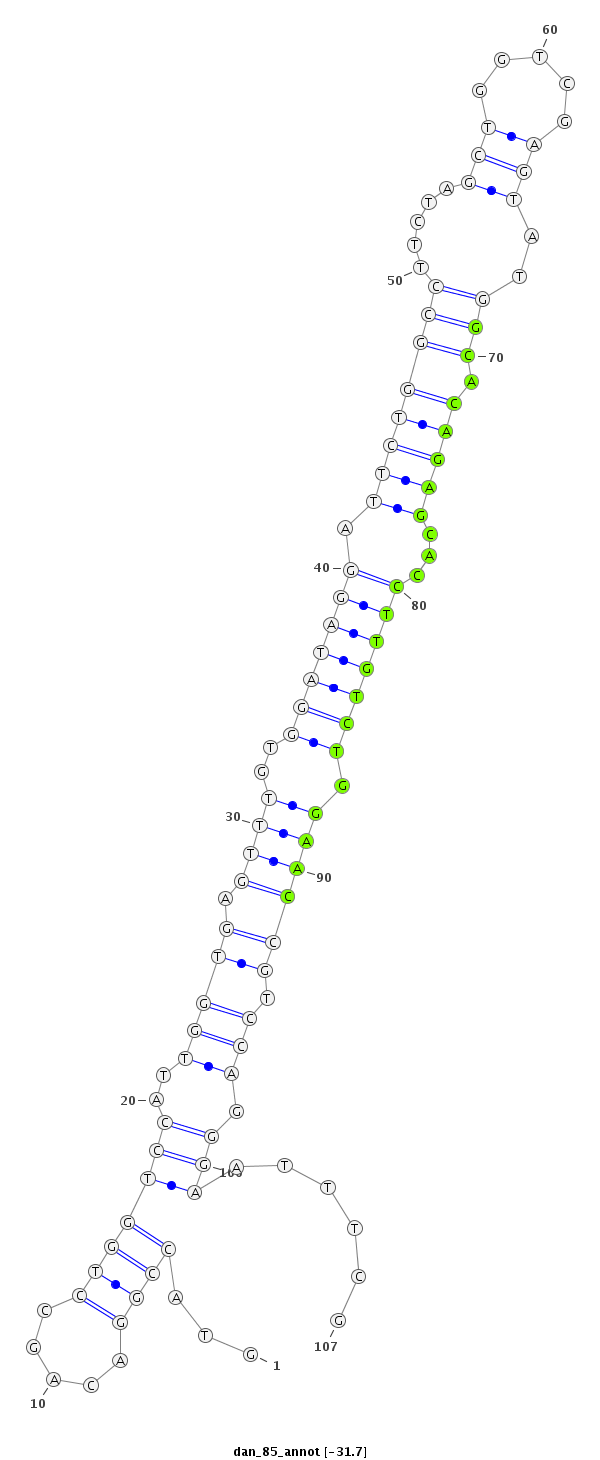
ID:dan_85 |
Coordinate:scaffold_13417:1620961-1621037 + |
Confidence:candidate |
Class:Canonical miRNA |
Genomic Locale:intergenic |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -32.5 | -31.8 | -31.7 | -31.7 |
 |
 |
 |
 |
intergenic
No Repeatable elements found
|
CCCCCCCTGAGTTCCTGCCCACATGGATCTAAAGCGTACCGGACAGCCTGGTCCATTGGTGAGTTTGTGGATAGGATTCTGGCCTTCTAGCTGGTCGAGTATGGCACAGAGCACCTTGTCTGGAACCGTCCAGGGAATTTCGAAGGAGATGCGGAAGTGAAGCGAGGGAGTGTGGAG
***********************************....((..........(((.((((((.((((..(((((((.((((((((.....(((.....)))..))).)))))...))))))).)))))).)))))))....))*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V105 male body |
V055 head |
V106 head |
M044 female body |
V039 embryo |
|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................GCACAGAGCACCTTGTCTGGAAC................................................... | 23 | 0 | 1 | 35.00 | 35 | 31 | 1 | 1 | 2 | 0 |
| ........................................................................................................CGCAGAGCACCTTGTCTGGAA.................................................... | 21 | 1 | 1 | 18.00 | 18 | 17 | 0 | 1 | 0 | 0 |
| .......................................................................................................GCGCAGAGCACCTTGTCTGGAA.................................................... | 22 | 1 | 1 | 13.00 | 13 | 12 | 1 | 0 | 0 | 0 |
| ........................................................................................................CGCAGAGCACCTTGTCTGGAAC................................................... | 22 | 1 | 1 | 10.00 | 10 | 10 | 0 | 0 | 0 | 0 |
| .......................................................................................................GCGCAGAGCACCTTGTCTGGAAC................................................... | 23 | 1 | 1 | 7.00 | 7 | 6 | 0 | 1 | 0 | 0 |
| ............................CTAAAGCGTACCGGACAGCC................................................................................................................................. | 20 | 0 | 1 | 4.00 | 4 | 4 | 0 | 0 | 0 | 0 |
| ........................................................................................................CGCAGAGCACCTTGTCTGG...................................................... | 19 | 1 | 1 | 4.00 | 4 | 4 | 0 | 0 | 0 | 0 |
| .......................................................................................................GCACAGAGCACCTTGTCTGGAA.................................................... | 22 | 0 | 1 | 4.00 | 4 | 3 | 1 | 0 | 0 | 0 |
| ........................................................................................................CGCAGAGCACCTTGTCTGGA..................................................... | 20 | 1 | 1 | 4.00 | 4 | 4 | 0 | 0 | 0 | 0 |
| ..................................................................................CCTTCTAGCTGGTCGAGTATG.......................................................................... | 21 | 0 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 |
| ............................CTAAAGCGTACCGGACAGCCT................................................................................................................................ | 21 | 0 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 |
| .........................................................................................................GCAGAGCACCTTGTCTGGAAC................................................... | 21 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| ........................................................................................................CGCAGAGCACCTTGTCTGGAATT.................................................. | 23 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................CTAAAGCGTACCGGACATCCT................................................................................................................................ | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| .......................................................................................................GCACAGAGTACCTAGCCTGGAACC.................................................. | 24 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................CGCATAGCACCTTGTCTGGAAC................................................... | 22 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................GCAGAGCACCTTGTCTGGAACC.................................................. | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................TCTAAAGCGTACCGGACAGCC................................................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................GCACAGAGCACCTTGTCTGGAACC.................................................. | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................CTAAAGCGTACCGGACAGT.................................................................................................................................. | 19 | 1 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 |
| ............................................................................................GGTCGAGTATGGCACAGAGC................................................................. | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................CCTTCTAGCTCGTCGAGTATG.......................................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................GGTGATGCGAGGAAGTGTGGAG | 22 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................CGCAGAGCACCTTGTCTGGG..................................................... | 20 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................GGTCGAGTATGGCACAGAGCAC............................................................... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................GCGCAGAGCACCTTGTCTGG...................................................... | 20 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................................CGCAGAGCACATTGTCTGGAAC................................................... | 22 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................CACAGAGCACCTTGTCTGGAA.................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................CTAAAGCGTACCGGACAGC.................................................................................................................................. | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................CCTTATAGCTGGCCGAGTATG.......................................................................... | 21 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................CGGCAGTGAAGCTAGGGAGTT..... | 21 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................GGCAGAGCACCTTGTCTGGAAT................................................... | 22 | 3 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................................CGCAGAGCACCTTGTCTGTAA.................................................... | 21 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................GTCCATTGGTGATTTTGT............................................................................................................. | 18 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 |
| ........................................TGACAGCGTGGTTCATTGGT..................................................................................................................... | 20 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................GATCTACTGCGTAGCGGAC..................................................................................................................................... | 19 | 3 | 9 | 0.33 | 3 | 0 | 0 | 0 | 3 | 0 |
| ................................................................TTGTGGATAGGATAGTGG............................................................................................... | 18 | 2 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................CTAAATCGTACCGGTCAAC.................................................................................................................................. | 19 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 1 | 0 |
| .....................................................CAGTGGTGAGGTTGTGGAAA........................................................................................................ | 20 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 1 | 0 |
| ............................................................................................................................................................GTGAAGCGAGATAGTGTGG.. | 19 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
|
GGGGGGGACTCAAGGACGGGTGTACCTAGATTTCGCATGGCCTGTCGGACCAGGTAACCACTCAAACACCTATCCTAAGACCGGAAGATCGACCAGCTCATACCGTGTCTCGTGGAACAGACCTTGGCAGGTCCCTTAAAGCTTCCTCTACGCCTTCACTTCGCTCCCTCACACCTC
***********************************....((..........(((.((((((.((((..(((((((.((((((((.....(((.....)))..))).)))))...))))))).)))))).)))))))....))*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M044 female body |
V106 head |
V055 head |
M058 embryo |
V105 male body |
V039 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................CACCTATCCTAAGAGTGGA............................................................................................ | 19 | 2 | 2 | 3.00 | 6 | 6 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................CCGTGTTTTGTGGTACAGAC....................................................... | 20 | 3 | 5 | 1.60 | 8 | 0 | 5 | 2 | 0 | 1 | 0 |
| .................................................................CCACCTATCCTAAGAGTGGA............................................................................................ | 20 | 3 | 3 | 1.00 | 3 | 2 | 0 | 0 | 1 | 0 | 0 |
| .......................................................................................................CGTGTTTTGTGGTACAGAC....................................................... | 19 | 3 | 20 | 0.75 | 15 | 10 | 4 | 0 | 1 | 0 | 0 |
| ......................................................................................................CCGTGTTTTGTGGTACAGA........................................................ | 19 | 3 | 20 | 0.50 | 10 | 10 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................CACCTATCCTAAGAGTGG............................................................................................. | 18 | 2 | 4 | 0.50 | 2 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................................GGCCTGTAGGGCCAGGTA......................................................................................................................... | 18 | 2 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .............................................................TAAGCCACCTATCCTAAGA................................................................................................. | 19 | 3 | 20 | 0.30 | 6 | 4 | 0 | 0 | 1 | 0 | 1 |
| ..................................................................CACCTATCCTAAGAGTGGAT........................................................................................... | 20 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................ACCTATCCTAAGAGTGGA............................................................................................ | 18 | 2 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................TGGCCTATAGGGCCAGGTA......................................................................................................................... | 19 | 3 | 8 | 0.13 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................................................CCACCTATCCTAAGAGTGG............................................................................................. | 19 | 3 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................GGAAGTTCGACCAGTTCA............................................................................. | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| Species | Coordinate | ID | Type | Alignment |
|---|---|---|---|---|
| droAna3 | scaffold_13417:1620911-1621087 + | dan_85 | candidate | CCCCCCCTGAGTTCCTGCCCACATGGATCTAAAG-----CGTACCG-----GACAGCCTGGTCCATTGGTGAGTTTGTGGATAG-GATTCTGGCCTTCTAGCTGGTCGAGTATGGCACAGAGCACCTTGTCTGGAACCGTCCAGGG---AATTTCGAAGGAGATGCGGAAGTGAAGCGAGGGAGT-GTGGA--------------------G |
| droBip1 | scf7180000396805:6932-7095 + | CGTCTCCCCAATTTCTGAGAACATGG-------CTAAATCCTTTCA-----GGCAACAGGGCCCCTTGGTGTGTATGTGGACCGCCACTCTGGTTTTCTAGCTCATCAAGTATAGCTCCTAGGACCTTGTCTGGAAGCGTGGAGGACGAAACTACGCAGAAGACGGGGACGTGCAG------------------------------------ | ||
| droTak1 | scf7180000414450:137216-137405 + | CACTCACTG--T-------CACATGGATCTAGGT-----GATACCGTTCCACG----ACACTCCACTTGTCGGTCACAAGATCGGGATTCTGCATCTCCAGCAGGTCGAAAACAGAGCACAAGGCTTCTCCCGGAATGACCAATGG---CAACACGAATGAGCACCGTATA-AGATCGAAAAAGTTGATGAATGTCTGTGCCATATGGGGGG |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
Generated: 10/20/2015 at 06:55 PM