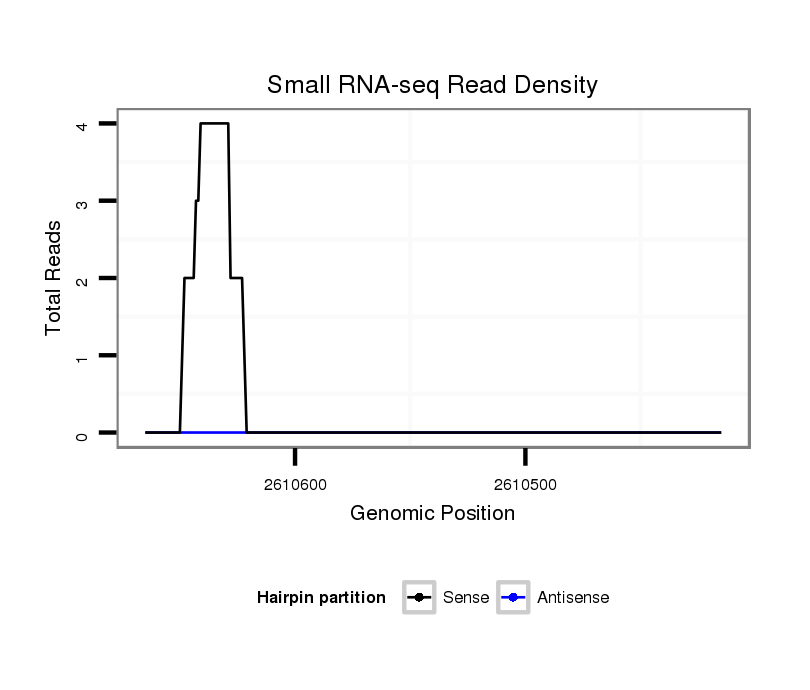
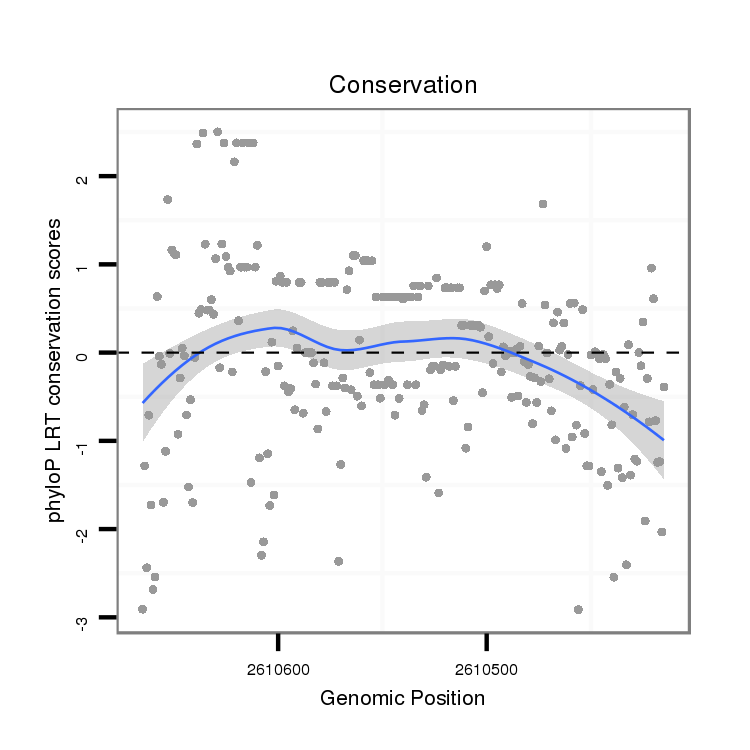
| droAna3 |
scaffold_12929:2610415-2610665 - |
dan_839 |
AAGCA---CTAATT-------CAAAAACTAT--AAACAAAAA-----------TGCAGCAGAAAGTTGAAGAGGTGAGTG--GGGAAAG---------G--GT----------------------ATTTTTTTAAAAAACAA--ATATAATTTTTGAGAAAAA---CAG----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATATTTTAGATTCTAGAAT-CTAAATTTTTAAAACCAAATATTAGTAATTTTTTTT---TGGA------AAATAGATCTTATTACTTTGTTTTCTATTCCTGTTTTTTTTTTTATTTTCGCTACGCCC----------CCGCCCCC--------GCCCCCC-------TTATTTTTACTA |
| droBip1 |
scf7180000395540:19515-19812 - |
|
AAGCA---CTAATAAAATAAGAA-AAATTAA--CCAACAAAA-----------TGCAGCAGAAAGTTGAAGAGGTGAGTG--AAGAAACTTGAT-A------------------------ATCTA-----------------------------AAA----AATTA--TATATTTCAAAACAAT---ATTTTTTATAAATTTTCAAAAATCTATATTTTTTTTCAAAATCTTGTTTAATATTTTTAAG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCCAGTTATTAATAATTGCTTTTAAATGGAGAGTTTAAATAGATCTCATTATTTTGTTCTCTATTCCTGTTCTTTTTTTTTTTGCTACGCCCCCG----------CCTTTAGGCCCTC---CCCCCT-TGGTAT-TTTTTTTTACTA |
| droKik1 |
scf7180000302696:10117-10272 + |
|
TCGCC---ACAGAT-------AAAAAGGATT--CACGGAAAA-----------TGCAGCAGAAAGTAGAAGAGGTAAGTA--ACTGTA------------------------AATTGTTGATGTA-----------------------------CAAGA-----------------------ATTTTAGCA-----------------AAGAATTA----------------------------TTGGGCAGGATAT-------------------------------------------------------------------------------------------------------------------------------TGTTA--TTTT--------GAAACG-----------------------------------------------------------------CAGGCCCCAA----------------------------------------------------------------------------------------A-AACTAATTAATGTTCATTT |
| droFic1 |
scf7180000453882:124612-124718 + |
|
AAT---------------------AAACTTT--TAAACAAAA-----------TGCAACAAAAAGTTGAAGAGGTGAGTC--TACGTGATTGGA-TAAGTGTT----------------------ATTTCTTTAAA----TGTTCTAACATTTTGGTAAATAA---AAG------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATAT |
| droEle1 |
scf7180000490546:415477-415634 + |
|
CCAAC---GCAAAA-------T--AAACTTT--TGATCAAAA-----------TGCAGCAGAAAGTTGAAGAGGTTAGTA--AATTCT-------------------------------------ACTACCAAAAC----TATTTTAAAATCTCAGTGAATAATTGCAG------------------------------------------------------------------------------------------------ATATTCAAAATGAATATATATTAA-----------------------------------------------------------------------------------------------AAT--TT------------GAAATG-------------------------------------------------------------------------------------------------------------------------------------------------------ATAT---CCAAAT--------TCTTATTTATTG |
| droRho1 |
scf7180000777786:3932-3985 + |
|
CAACC---GCAAAA-------C--AAATTTT--TAAACAAAA-----------TGCAGCAAAAAGTTGAAGAGGTTAGT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droBia1 |
scf7180000302075:1181108-1181263 + |
|
ACGCA---AT---A-------T--AAACTTTTACAAACAAAA-----------TGCAGCAGAAAGTTGAAGAGGTGAGTG--CCTTCACTTGGG-A----------------------------------------------------------------AAAT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTGTTAGAGAT--------TAGTATACTTTTT--TTTTAATTC-----TAAG-----------------------------T--TTAGAATTACACATAAGAAA----------------GCTTTAA----------------------------------------------------------------------------------------A-AAATATTTTTTTTTATTCT |
| droTak1 |
scf7180000415659:152902-153118 - |
|
AAGCGCACGGAAAA-------T--AAACTTT--TTAGCAAAA-----------TGCAGCAGAAAGTTGAAGAGGTTAGTA--CTTTTACTTGGA-T------------------------C---------------------------------TATGGATAATTT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TATAGATTT--------TG--------------------TTCATGAAAATACAGATAATATTTAATTTTCACTCCTTAATATTAGCAA--AAA-------------------------------------------AATTATCTTTTATTTTTAA---TTTGATGTACCT------------ATACCATTTTTTATA------------CAAAA-CAATATTTTTATTTACTTT |
| droEug1 |
scf7180000409110:768932-769038 + |
|
TTATT---TTTGTG-------TA------------------A-----------TATTCTAGT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTGTTG------------------------------------CTG--------------------------------------------------------------------------------------------------------------------CGTTTTGGTCCATGTTGTTGCTTTTTCTCCTCGT------------TCTGCTCTTCCACCTTCTCTTTT---GCGTTC--------CTCCCTTTTCCT |
| dm3 |
chrX:12391581-12391632 + |
|
CAGCA---AA---C-------TA-AAACTTT--TCATCAAAA-----------TGCAGCAGAAAGTCGAAGAGGTTAGT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droSim2 |
x:11749358-11749630 + |
|
AAGCA---TTAAAA-------A---GACATT--A----AGTAGGAACTTAAAAGTTCGCAAAAACTAGAGCATTTTATATTTTATGACTGAG-GAAAAGTCTTATAACACATATTCGTTAATGTT-----------------------------GAACA-----------------------ATAACATTT-----------------TACGATTA----------------------------TAGTTTACGAAATAATCGTTAATATCCAACAGATGATTACATTAGTTTTGCAGTTCATGAAG----------------------------TTGTTC------------------------------------TTG--------------------------------------------------------------------------------------------------------------------TATTTTCTCTTATTTTTTGGTTTTTGATTTTTCT------------CCACCCATTCTCCTATCTCC---TTTGCTTCC--------CTGCATTCTCCT |
| droSec2 |
scaffold_21:148319-148567 + |
|
GAACT---TAAAAA-------T---------------------------------TCGCAAAAACTAGAGCATTTTATATTTTATGACTGAG-GAAAAGTCTTATAACATATATTCGTTAATGTT-----------------------------GAACA-----------------------ATAACAGTT-----------------TACGATTA----------------------------TAGTTTACGAAATAATCGTTAATATCCAACAGATGATTACATTAGTTTTGCAGTACATGAAG----------------------------TTGTTC------------------------------------TTG--------------------------------------------------------------------------------------------------------------------TATTTTCTCTTATTTTTTGGTTTTTGATTTTCCT------------CCACCCATTCTCCTATCTCC---TTTGCTTCC--------CTGCATTCTCCT |
| droYak3 |
X:6666857-6667002 + |
|
AAACA---GCAAAG-------TA-AAACTAC--TCGTCAAAA-----------TGCAGCAGAAAGTTGAAGAGGTAAGTA--AACATATAAATC-T----------------------TAGCA------GTA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGCTAATATATATTTTTTGATTCTAACTTGGCTAAAAAGATAATA----------CTATTTTTTATTT---AT---------------TGAATAAAT |
| droEre2 |
scaffold_4690:14467475-14467529 - |
|
CCACA---GCAAAG-------TA-AAAAGTT--TCGTCAAAA-----------TGCAGCAGAAAGTTGAAGAGGTTAGT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| dp5 |
XL_group1a:3133697-3133798 + |
|
TTTTT---TTTTAC-------T----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGAAAGAGCCTTCTATTTGGGTCTGTTTTCCTGGTTTCATTATGATCGGGATGATCCTT----------CCGTTTGTGTCTC---TCGATC--------CTGTTTTTAATG |
| droPer2 |
scaffold_12:144352-144453 - |
|
TTTTT---TTTTAC-------T----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGAAAGAGCCTTCTATTTGGGTCTGTTTTCCTGGTTTCATTATGATCGGGATGATCCTT----------CCGTTTGTGTCTC---TCGATC--------CTGTTTTCATGG |
| droWil2 |
scf2_1100000004590:4181339-4181461 + |
|
AAAAA---CCAAAA-------G--AAACTT------CAAATA-----------TGCAGCAAAAGGTTGAAGAGGTTAGTA--TCAACACTAAGG-G----------------------AAATA------CTGT---------------------GTGAA--------------------ACTAT---ATTT-----------------AAAGATTG----------------------------T---------------------------------------------------------------------------------------------T-----------------------------------------------------------ACTAAGAAATGACAAATAAT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTGGT |
| droVir3 |
scaffold_12963:7741677-7741756 - |
|
TTTTA---GAAATT-------TA-T-----------------------------------------------------TG--TTAATAA----------------------------------------------------------------------------------------------------------------------------------TTTTGAAAATGTAA---------------------------------------------------------------------------------------------------------TT------------------------------------TTGTATTTATT--TTCT------------TCTG-------------------------------------------------------------------------------------------------------------------------------------------------------TTGT---TTTTTT--------TTTTTTTTTTGC |
| droMoj3 |
scaffold_6328:1901053-1901275 + |
|
AAACC---GAATAA-------A--AAA--AA--TACACAAAA-----------TGCAGCAAAAAGCCGATGAGGTGAGTA--GCAAAATAGG--------------------------AAATAGA-----------------------------ACAAA--------------------ATAAT---AGCAT----AA----------AACGATCG----------------------------CAGCA-----TGT-----------------------------------------------------TGCGACGGCGACTGCTGCTGTTGTCCTCTGCAACATCCTGCCCAACTTTGTTGCAGAACTTTAATTCTTTCGTTTTT--GTCT------------TTCG------------------------------------------------------------------------------------------------------------------------------------CTATGCGTT---GAAC---TCTCTTTG---CT--------CTCCTTTACTAT |
| droGri2 |
scaffold_14853:7611169-7611354 - |
|
AACAA---AGAACAACAACA-TATAAATCATAATCGACAGAA-----------TGCAGCAAAAAGCCGATGAGGTAAGTG--TAATAAATAC---------------------------------------------------------------------------GC------------------------------------------------------------------------------------------------ATAGACAAAATAGATA---------------GATCGTTGAGTTGTTGCTGCAACG--------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTTGTCGCTGGGCACACATCCTGTGCAACATTGTTGCT------------GAACTCGCATTTCTTTCGCC---TTTG-------------------CTGGGC |