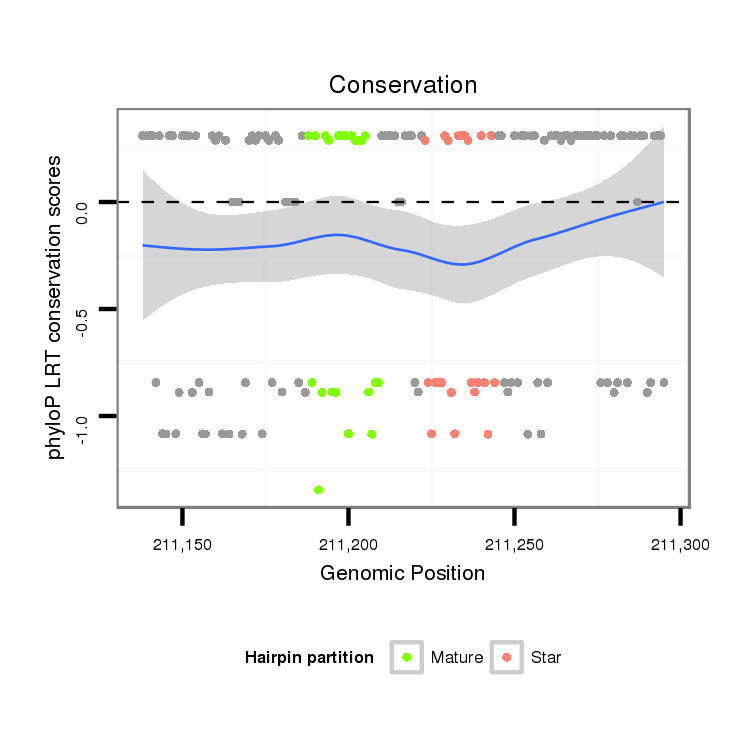
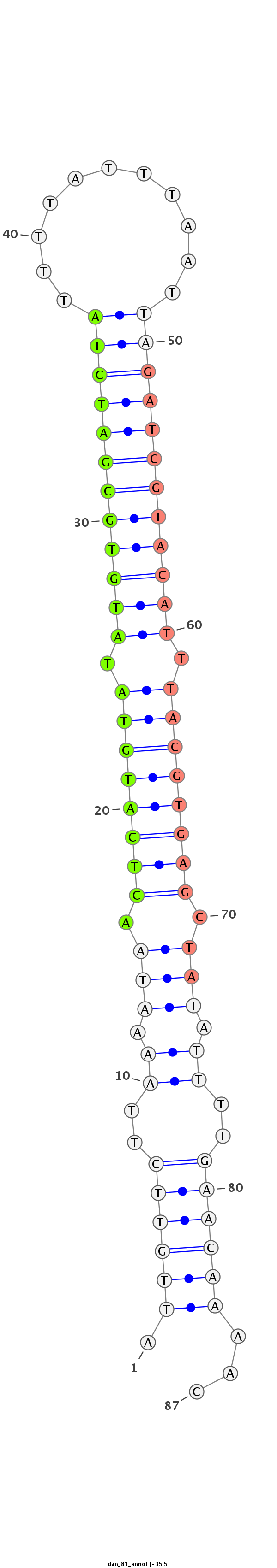
ID:dan_81 |
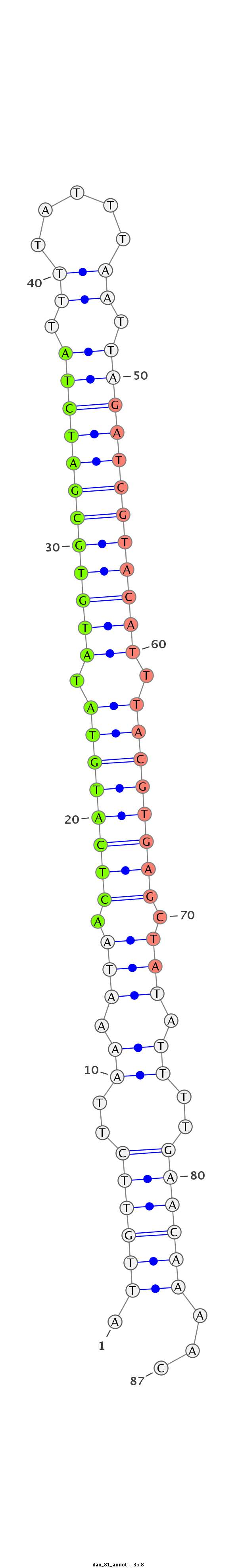
Coordinate:scaffold_13335:211188-211245 + |
Confidence:confident |
Class:Testes-restricted |
Genomic Locale:intergenic |
[View on UCSC Genome Browser {Cornell Mirror}] |
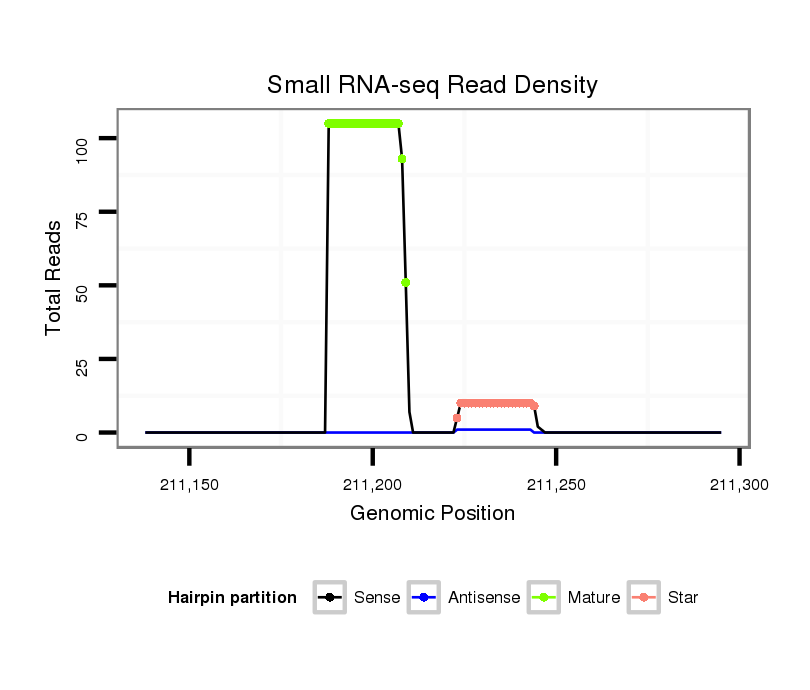
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -35.5 | -34.8 |
 |
 |
intergenic
No Repeatable elements found
|
AATTGTGAAATATAAATATATTCTTCAAATCGGAGATTGTTCTTAAAATAACTCATGTATATGTGCGATCTATTTTATTTAATTAGATCGTACATTTACGTGAGCTATATTTTGAACAAAACTTAACAACAAATTTAATTCAACAATATTAAATTTTC
***********************************.((((((..((.(((.((((((((.((((((((((((.((.....)).)))))))))))).)))))))).))).))..))))))...************************************ |
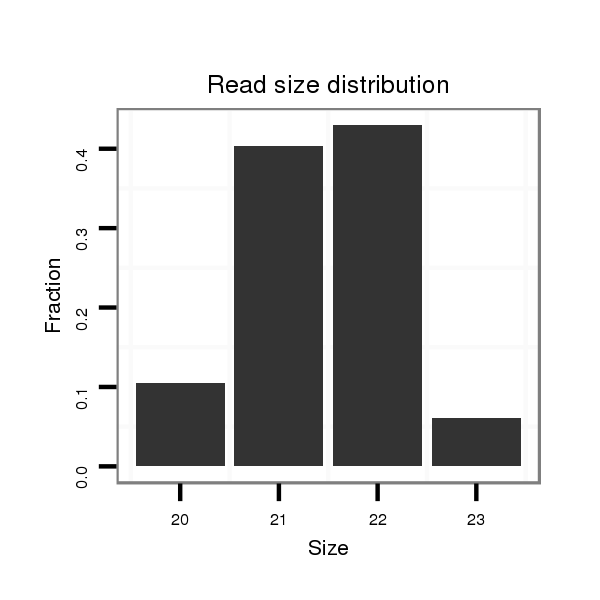
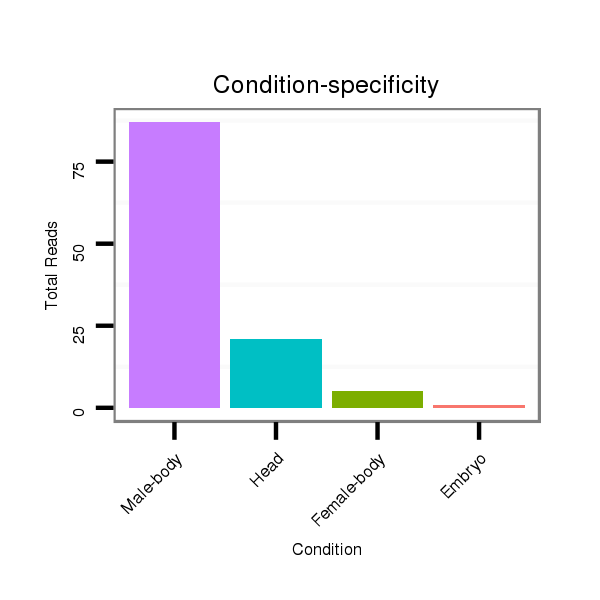
Read size | # Mismatch | Hit Count | Total Norm | Total | V105 male body |
V055 head |
M044 female body |
M058 embryo |
V106 head |
V039 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................ACTCATGTATATGTGCGATCTA...................................................................................... | 22 | 0 | 1 | 44.00 | 44 | 35 | 6 | 1 | 0 | 2 | 0 |
| ..................................................ACTCATGTATATGTGCGATCT....................................................................................... | 21 | 0 | 1 | 42.00 | 42 | 31 | 9 | 1 | 1 | 0 | 0 |
| ..................................................ACTCATGTATATGTGCGATC........................................................................................ | 20 | 0 | 1 | 12.00 | 12 | 11 | 1 | 0 | 0 | 0 | 0 |
| ..................................................ACTCATGTATATGTGCGATCTAT..................................................................................... | 23 | 0 | 1 | 7.00 | 7 | 5 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................................GATCGTACATTTACGTGAGCTA................................................... | 22 | 0 | 1 | 4.00 | 4 | 3 | 0 | 1 | 0 | 0 | 0 |
| ..................................................ACTCATGTATATGTGCGATCTAA..................................................................................... | 23 | 1 | 1 | 4.00 | 4 | 4 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................ATCGTACATTTACGTGAGCTA................................................... | 21 | 0 | 1 | 3.00 | 3 | 2 | 1 | 0 | 0 | 0 | 0 |
| ..................................................ACTTATGTATATGTGCGATCT....................................................................................... | 21 | 1 | 1 | 2.00 | 2 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..................................................AATCATGTATATGTGCGATCT....................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................GATCGTACATTTACGTGCGCTAT.................................................. | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................ACTCATGTATATTTGCGATCTA...................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................ACTCATGTATATATGCGATCTA...................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................ATCGTACATTTACGTGAGCTATA................................................. | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................ACTCATGGATATGTGCGATCT....................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................GCTCATGTATATGTGCGCTCT....................................................................................... | 21 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................ACGCATGTATATGTGCGATCT....................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................ACTCATGGATATGTGCGATCTA...................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................ACTCATGTATATGTACGATCTC...................................................................................... | 22 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................ACTCATGTATATGTGCGATCTATA.................................................................................... | 24 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................AGTCATGTATATGTGCGATCTA...................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................ACTAATGTATATGTGCGATCTAA..................................................................................... | 23 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................ATCGTACATTTACGTGAGCTAT.................................................. | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................ACTCATGTATATGTGCGATAT....................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................ACTCATGTATATGTGCGATCC....................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................ACTCATGTGTATGTGCGATCT....................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................GATCGTACATTTACGTGAGCT.................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..................................................ACTCATGTATATGAGCGGTCTA...................................................................................... | 22 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................GATCGTACATTTACGTGAGCGA................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................ACTCATGTATATGTGCGTGCTG...................................................................................... | 22 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................ACTCATGTATATGTGCTATCTA...................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................ATGTATATGTGCGATCTAAA.................................................................................... | 20 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................ACTCGTGTGTATGTGCGCTCTA...................................................................................... | 22 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................AAACAAAACTTAACAACAA.......................... | 19 | 1 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..........AATAAATATATGCTTCAAACC............................................................................................................................... | 21 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
|
TTAACACTTTATATTTATATAAGAAGTTTAGCCTCTAACAAGAATTTTATTGAGTACATATACACGCTAGATAAAATAAATTAATCTAGCATGTAAATGCACTCGATATAAAACTTGTTTTGAATTGTTGTTTAAATTAAGTTGTTATAATTTAAAAG
************************************.((((((..((.(((.((((((((.((((((((((((.((.....)).)))))))))))).)))))))).))).))..))))))...*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V106 head |
M044 female body |
M058 embryo |
|---|---|---|---|---|---|---|---|---|
| .....................................................................................CTAGCATGTAAATGCACTCGA.................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 |
| .............................................................................................................AAAAGTTGTTTTGAATGGTTGT........................... | 22 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ...ACACTTTATAGTTATATAA........................................................................................................................................ | 19 | 1 | 2 | 0.50 | 1 | 0 | 0 | 1 |
| ..........................................................................................ATGGAAATGTACTCGATA.................................................. | 18 | 2 | 6 | 0.17 | 1 | 1 | 0 | 0 |
| .............................................................CCACGGTAGATAAAATGAA.............................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| Species | Coordinate | ID | Type | Alignment |
|---|---|---|---|---|
| droAna3 | scaffold_13335:211138-211295 + | dan_81 | confident | AATTGTGAAATATAAATATATTCTTCAAATCGGAGATTGTTCTTAAAATAACTCATGTATATGTGCGATCTATTTTATTTAATTAGATCGTACATTTACGTGAGCTAT---------------ATTTTGAACAAAACTTAACAACAAATTTAATTCAACAATATTAAATTTTC |
| droBip1 | scf7180000396431:1453190-1453352 + | AATTATTCAAGTTAATTGGCATCTGCC---AAGAGAGTGCTCA----GTTATTGTTGATTATTTGCGAAACGTTTTA--TAACAAGGGTACACTGTTACAAAAAATGTAAAACATTTAAATTAACACTAAAAAAGCCCTAACAACAAATTTAACTTATTAACAT-AATCTTTT |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
Generated: 10/20/2015 at 06:50 PM