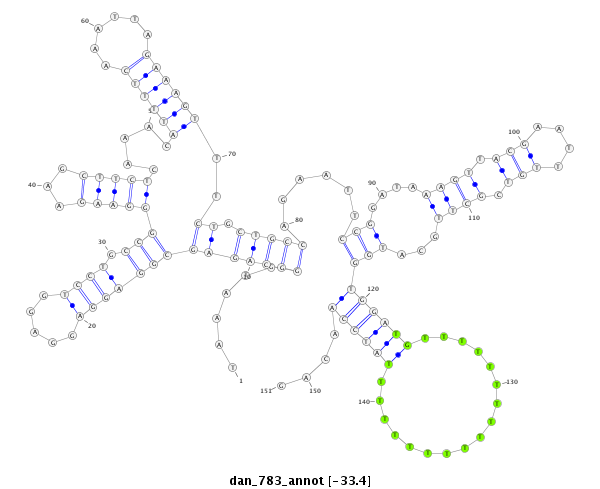
| ...........................................................................................................................................................GAGCGACCGTAGCACCTACAAA.......................................................................... |
22 |
3 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
| .....................................................................................................................................................................CCCACCTACAAAAAGAAAAAAA................................................................ |
22 |
2 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................................................ACCACCCACAAAAAAAAA.................................................................... |
18 |
1 |
20 |
0.20 |
4 |
4 |
0 |
0 |
0 |
0 |
| ......................................................................................................................................................................CCACCTCCAAAAAAAAAAAA................................................................. |
20 |
1 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................................................ACCACCCACAAAAAAAAAA................................................................... |
19 |
1 |
16 |
0.19 |
3 |
3 |
0 |
0 |
0 |
0 |
| .......................................................................................................................................................................CACCTACAGAAAAAAAAAA................................................................. |
19 |
1 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................................................ACCACCCACAAAAAAAAAGAAAAA.............................................................. |
24 |
2 |
7 |
0.14 |
1 |
1 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................................................ACCACCTCCAACAAAACAAAAAA............................................................... |
23 |
3 |
7 |
0.14 |
1 |
1 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................................................................................................................GTGAGATCCCCCTTTACCGA.. |
20 |
3 |
7 |
0.14 |
1 |
0 |
0 |
1 |
0 |
0 |
| .....................................................................................................................................................................ACCACCCACAAAACAAGAAAAAAAA............................................................. |
25 |
3 |
9 |
0.11 |
1 |
1 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................................................ACCACCTCCAACAAAAAAAA.................................................................. |
20 |
2 |
9 |
0.11 |
1 |
1 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................................................ACCACCCACAAAAAAAAAAAAGA............................................................... |
23 |
2 |
10 |
0.10 |
1 |
1 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................................................ACCACCCACAACAAAAAAAAA................................................................. |
21 |
2 |
20 |
0.10 |
2 |
2 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................................................ACCACCCACAAGAAAAAAAA.................................................................. |
20 |
2 |
20 |
0.10 |
2 |
1 |
0 |
1 |
0 |
0 |
| ..................................................................................................................................................................................AAAAAAAAAAAAATAAGGTTGTCAT................................................ |
25 |
3 |
11 |
0.09 |
1 |
1 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................................................ACCACCAACAAAAAAAACACAAAAAA............................................................ |
26 |
3 |
11 |
0.09 |
1 |
1 |
0 |
0 |
0 |
0 |
| ......................................................................................................................................................................CCACCCACAAAAAAAAAAAAAA............................................................... |
22 |
1 |
12 |
0.08 |
1 |
1 |
0 |
0 |
0 |
0 |
| ......................................................................................................................................................................CCACCGACAAAAAAAAACAAAAAA............................................................. |
24 |
2 |
12 |
0.08 |
1 |
1 |
0 |
0 |
0 |
0 |
| ......................................................................................................................................................................CCACCAACAAAAAAAAAAAAA................................................................ |
21 |
1 |
13 |
0.08 |
1 |
1 |
0 |
0 |
0 |
0 |
| .........................................................................................................................................................................................AAAAAAAAAAGTTGTCCTGA.............................................. |
20 |
2 |
15 |
0.07 |
1 |
0 |
0 |
0 |
1 |
0 |
| .....................................................................................................................................................................ACCACCAACAAAAAAAAAAAACAACAA........................................................... |
27 |
3 |
16 |
0.06 |
1 |
1 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................................................................AAAAAAAAAAAAAGGTTGTCAT................................................ |
22 |
2 |
16 |
0.06 |
1 |
0 |
0 |
0 |
0 |
1 |
| ......................................................................................................................................................................CCACCAACAAAAAAAAAAAA................................................................. |
20 |
1 |
18 |
0.06 |
1 |
1 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................................................ACCACCTGCACAAAAAAACAA................................................................. |
21 |
3 |
19 |
0.05 |
1 |
0 |
0 |
1 |
0 |
0 |
| ......................................................................................................................................................................CCACCCACAAAAAAAAAAAAA................................................................ |
21 |
1 |
19 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
| ......................................................................................................................................................................CCACCCACAAAAACAAAAAAA................................................................ |
21 |
2 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
| ....................................................................................................................................................................................................................GATGGTCCTTATGGGTC...................... |
17 |
2 |
20 |
0.05 |
1 |
0 |
1 |
0 |
0 |
0 |
| ......................................................................................................................................................................CCACCTCCAAAAAAAACAAGA................................................................ |
21 |
3 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................TTCAAACGCTTAAATTAT................................................................................................................................................................................................................. |
18 |
2 |
20 |
0.05 |
1 |
0 |
0 |
1 |
0 |
0 |
| ......................................................................................................................................................................CCACCCACAAGAAAAAAAACAAA.............................................................. |
23 |
3 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
| ......................................................................................................................................................................................AAAAAAAAAATAGGTTGTTG................................................. |
20 |
2 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
| ......................................................................................................................................................................CCACCCACAAACAAAAAAAAAA............................................................... |
22 |
2 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................................................ACCACCCACAAGAAAAAAA................................................................... |
19 |
2 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
| ......................................................................................................................................................................CCACCCACAAAAAAAAAACAAAA.............................................................. |
23 |
2 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................................................ACCACCCACAAAAGAAAAAA.................................................................. |
20 |
2 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................................................ACCACCCACAAACAAAAAAA.................................................................. |
20 |
2 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................................................ACCACCTGCAACAAAAAA.................................................................... |
18 |
2 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
| .....................................................................................................................................................................ACCACCCACAAAAAGAAAAA.................................................................. |
20 |
2 |
20 |
0.05 |
1 |
0 |
0 |
1 |
0 |
0 |
| .....................................................................................................................................................................ACCACCCGCCAAAAAAAAAAAAAAAA............................................................ |
26 |
3 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |