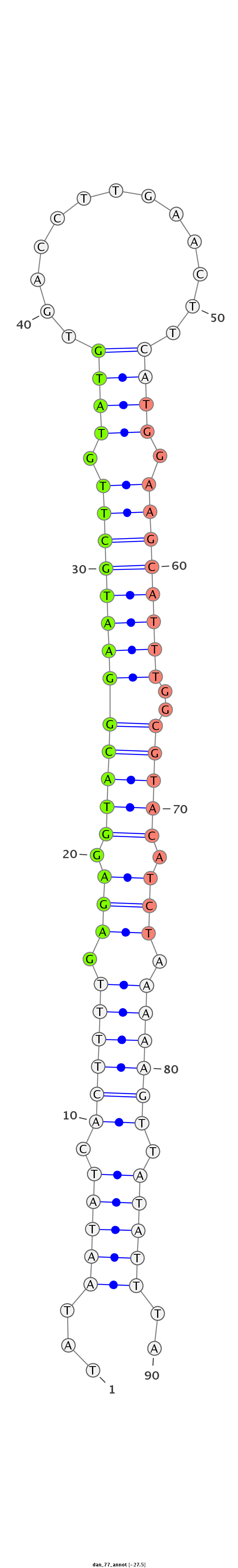
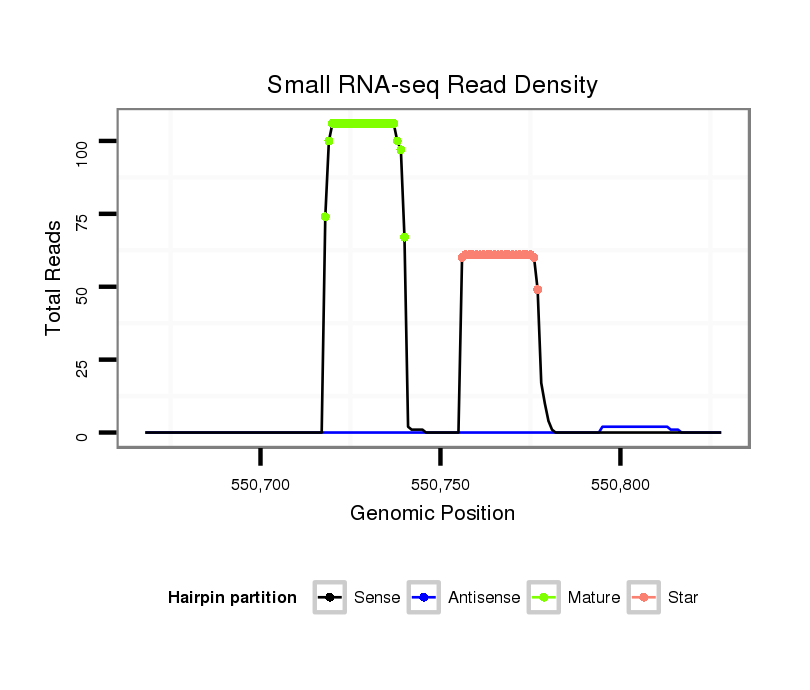
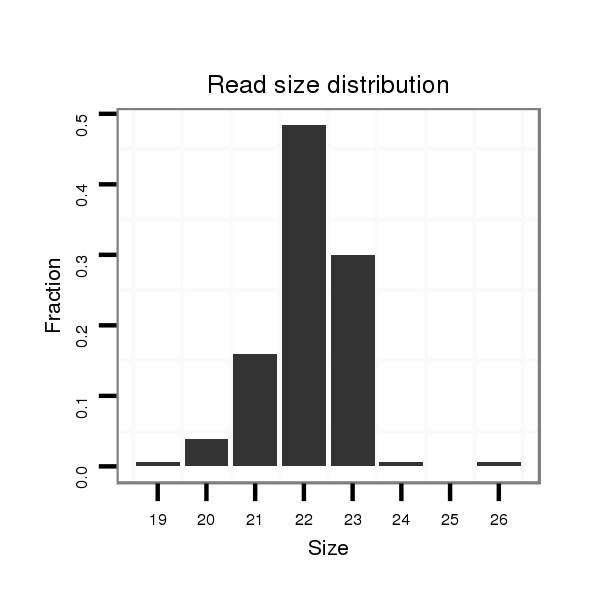
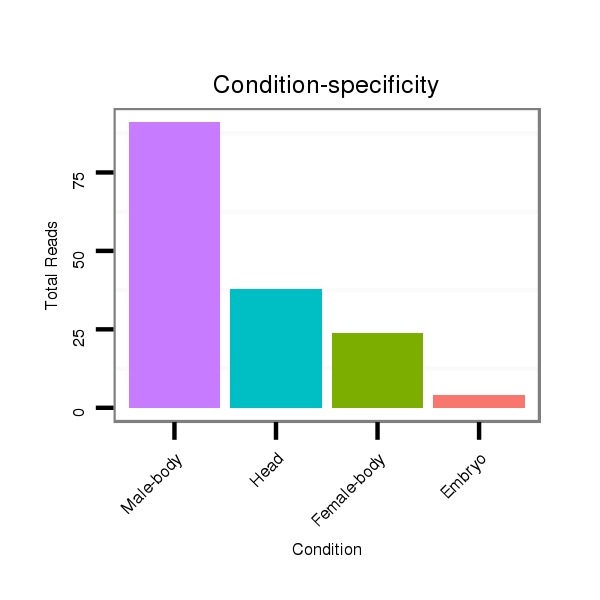
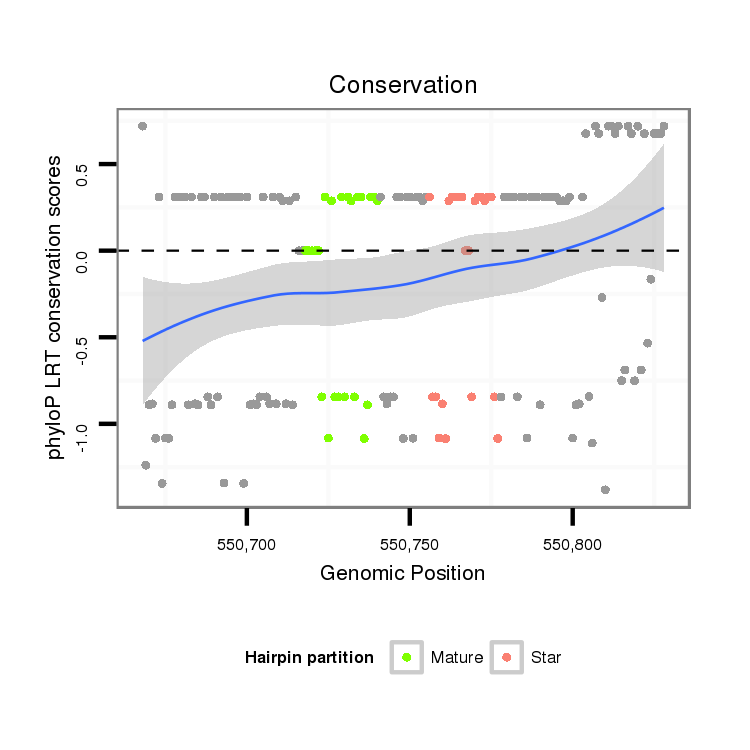
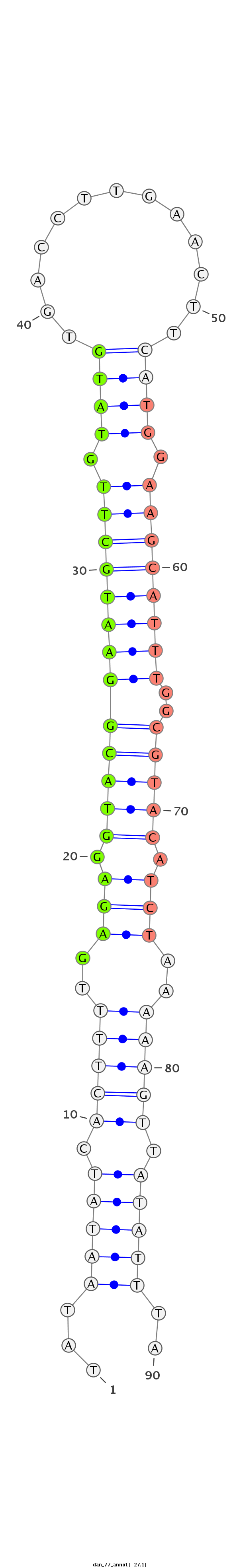
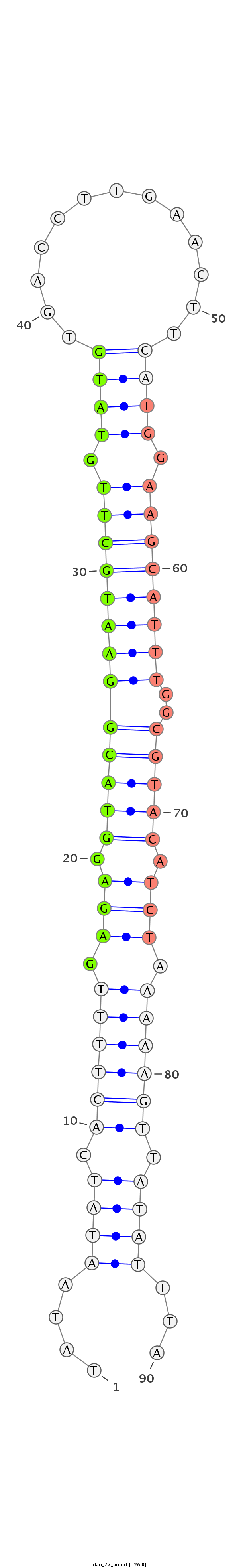
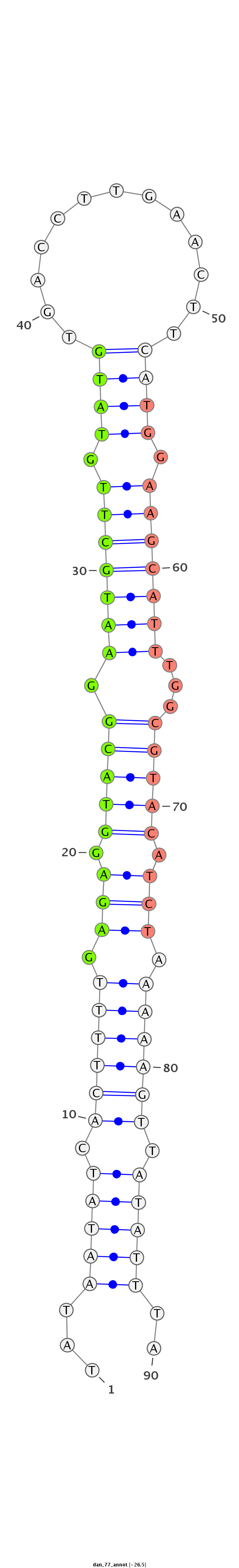
ID:dan_77 |
Coordinate:scaffold_12903:550718-550778 + |
Confidence:confident |
Class:Testes-restricted |
Genomic Locale:antisense_to_intron |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -27.1 | -26.8 | -26.5 |
 |
 |
 |
Antisense to intron [Dana\GF15947-in]
No Repeatable elements found
|
ACTATTCATTATAATAATATCTTATGAATATCTTATATAATATCACTTTTGAGAGGTACGGAATGCTTGTATGTGACCTTGAACTTCATGGAAGCATTTGGCGTACATCTAAAAAGTTATATTTATTTCCCAATATCCCTCAGAACTTTTCAAACCGCCCT
***********************************...(((((.((((((.(((.(((((((((((((.((((.............)))).))))))))..))))).))).)))))).)))))..************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | V105 male body |
M044 female body |
V106 head |
V055 head |
M058 embryo |
|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................GAGAGGTACGGAATGCTTGTATG........................................................................................ | 23 | 0 | 1 | 40.00 | 40 | 29 | 1 | 7 | 3 | 0 |
| ........................................................................................TGGAAGCATTTGGCGTACATCT................................................... | 22 | 0 | 1 | 31.00 | 31 | 11 | 15 | 0 | 4 | 1 |
| ..................................................GAGAGGTACGGAATGCTTGTAT......................................................................................... | 22 | 0 | 1 | 25.00 | 25 | 17 | 1 | 7 | 0 | 0 |
| ...................................................AGAGGTACGGAATGCTTGTATG........................................................................................ | 22 | 0 | 1 | 20.00 | 20 | 13 | 1 | 3 | 3 | 0 |
| ........................................................................................TGGAAGCATTTGGCGTACATC.................................................... | 21 | 0 | 1 | 11.00 | 11 | 5 | 2 | 0 | 1 | 3 |
| ........................................................................................TGGAAGCATTTGGCGTACATCTA.................................................. | 23 | 0 | 1 | 7.00 | 7 | 5 | 2 | 0 | 0 | 0 |
| ........................................................................................TGGAAGCATTTGGCGTACATCTAA................................................. | 24 | 0 | 1 | 6.00 | 6 | 4 | 2 | 0 | 0 | 0 |
| ...................................................AGAGGTACGGAATGCTTGTAT......................................................................................... | 21 | 0 | 1 | 5.00 | 5 | 1 | 0 | 3 | 1 | 0 |
| ....................................................GAGGTACGGAATGCTTGTATG........................................................................................ | 21 | 0 | 1 | 5.00 | 5 | 5 | 0 | 0 | 0 | 0 |
| ..................................................GAGAGGTACGGAATGCTTGT........................................................................................... | 20 | 0 | 1 | 5.00 | 5 | 2 | 0 | 2 | 1 | 0 |
| ........................................................................................TGGAAGCATTTGGCGTACATCTAAA................................................ | 25 | 0 | 1 | 3.00 | 3 | 0 | 0 | 1 | 0 | 2 |
| ..................................................GAGAGGTACGGAATGCTTGTA.......................................................................................... | 21 | 0 | 1 | 3.00 | 3 | 2 | 0 | 0 | 1 | 0 |
| ........................................................................................TGGAAGCATTTGGCGTACATCTT.................................................. | 23 | 1 | 1 | 2.00 | 2 | 0 | 1 | 0 | 1 | 0 |
| ...................................................AGAGGTACGGAATGCTTGT........................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................................GAGGCACGGAATGCTTGTATG........................................................................................ | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................TGGAAGCATTTGGCGTACATCTAT................................................. | 24 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................TGGAAGCATTTGGCGTACATCTAC................................................. | 24 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................TGGAAGCATTTGACGTACATCT................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................TGAAGCATTTGGCGTACATCT................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................................GAGTTACGGAATGCTTGTATG........................................................................................ | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................GAGAGGTACGGAATGCTTGTCTG........................................................................................ | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................GAGAGGTACGGAATGCTTGTATGT....................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| .........................................................................................GGAAGCATGTGGCGTACATCT................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................GAGAGGTACGGAATGTTTGTATG........................................................................................ | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................GAGGTACGGAATGCTTGTATGTGACC................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................GAGAGGTACGGAATGCTTGTATA........................................................................................ | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................GAGAGGTAAGCAATGCTTGTAT......................................................................................... | 22 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................CGTAAGCATTTGGCGTACATCT................................................... | 22 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................TTGAAGCATTTGGCGTACATCC................................................... | 22 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 |
| ........................................................................................TGGAAGCATTTGGCGTACATCTAAAA............................................... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................GAGAGGTATGGAATGCTTGTAT......................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................GAGAGGTACGGAATGCTTGTTTG........................................................................................ | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ........................................................................................TGGAAGCATTTGGCGTACATAT................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................GGAAGCATTTGGCGTACATCT................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................GGAGTGATCGTATGTGACCTTG................................................................................ | 22 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................TGGAAGCATTTGGCGTACACC.................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..................................................GAGAGGTACGGAATGCTTGTG.......................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................GAGAGGTACGGAATGCTCGTATG........................................................................................ | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..................................................GAGAGGTATGAAATGCTTGTCTG........................................................................................ | 23 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................TGGAAGCATTTGGCGTACAT..................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..................................................GAGAGGTACGCAATGCTTGTATG........................................................................................ | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................TGGAAGCATTTTGCGTACATC.................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................TCGCTTGTGAGAGGTACGG.................................................................................................... | 19 | 2 | 12 | 0.33 | 4 | 2 | 0 | 2 | 0 | 0 |
| ...........................................CGCTTGTGAGAGGTACGG.................................................................................................... | 18 | 2 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................................CTTGTGAGAGGTACGGGA.................................................................................................. | 18 | 2 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..........................................TCGCTTGTGAGAGGTACG..................................................................................................... | 18 | 2 | 14 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 |
|
TGATAAGTAATATTATTATAGAATACTTATAGAATATATTATAGTGAAAACTCTCCATGCCTTACGAACATACACTGGAACTTGAAGTACCTTCGTAAACCGCATGTAGATTTTTCAATATAAATAAAGGGTTATAGGGAGTCTTGAAAAGTTTGGCGGGA
************************************...(((((.((((((.(((.(((((((((((((.((((.............)))).))))))))..))))).))).)))))).)))))..*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M058 embryo |
V055 head |
M044 female body |
V039 embryo |
|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................................................AGGGTTATAGGGAGTCTTGAAA............ | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................AGGGTTATAGGGAGTCTTG............... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ........................................................................................................................TAAATATAGGGTTATCGAGAGT................... | 22 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ...........................................................................TGGAACTTGTAGTGCCTTCG.................................................................. | 20 | 2 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 |
| ..................................................CCCTCCATTCCTGACGAACA........................................................................................... | 20 | 3 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 |
| ..................................................................................................ACCGCATGTAGATGATACA............................................ | 19 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droAna3 | scaffold_12903:550668-550828 + | dan_77 | ACTATTCATTATAATAATATCTTATGAATATCTTATATAATATCACTTTTGAGAGGTACGGAATGCTTGTATGTGACCTTGAACTTCATGGAAGCATTTGGCGTACATCTAAAAAGTTATATTTATT-----------------------------------TCCCAATATCCCTCA---------------------------------------------GAACTTTTCAAACCGCCCT |
| droBip1 | scf7180000395912:19320-19551 + | AAATGTGCGAATAAAATAATTATGTCAATATGTATAGTGTTTTCTCAT-------ATCCAAAGTGTTTTAATGTATTTTTTAAATTCATAACTTCATTT--TGTACATTGGAAAAATTCTATATATTTAAACCTATGTTAAAGAGTTAAAGTTGAGTTGAGTTCCCACATTCATTCAATGTGATATTTCCAAATGGGGTATTAACATCTTGGCATTGGTATATAACTACTCTAGCTGCCCT | |
| droRho1 | scf7180000778091:424734-424760 + | AT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------CCATCC---------------------------------------------CAACTTTTCAAACCACCCT |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
Generated: 09/08/2015 at 08:01 PM