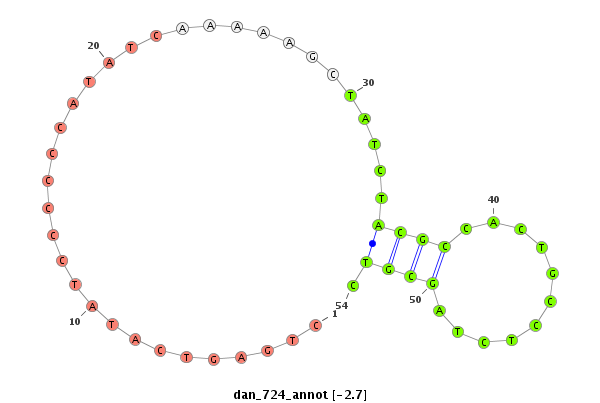



ID:dan_724 |
Coordinate:scaffold_12916:14848177-14848327 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -2.5 | -1.9 | -1.7 |
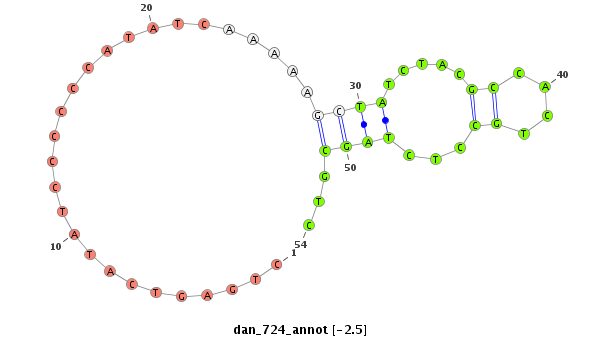 |
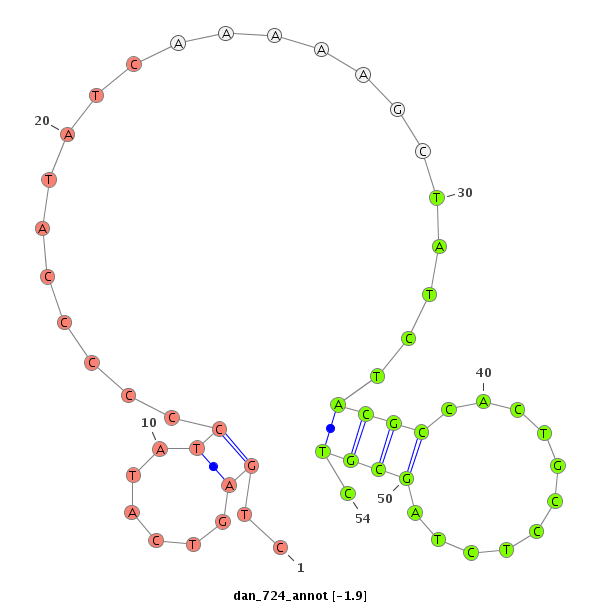 |
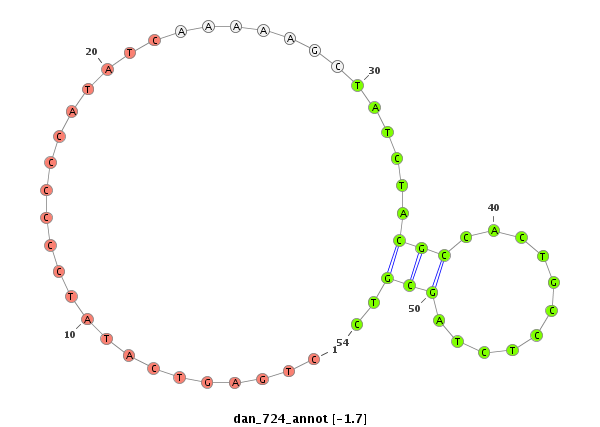 |
exon [dana_GLEANR_14808:2]; CDS [Dana\GF14047-cds]; intron [Dana\GF14047-in]; intron [Dana\GF14047-in]
No Repeatable elements found
| mature | star |
| -----------------------------#####################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- CAGTAATATTATTATTATTATTATTGCAGATTGCGGATTATTATCGCAGTGTAAAATAAAACTTACAAAAAAACAGGATATTATTCTGAGTCATATCCCCCCATATCAAAAAGCTATCTACGCCACTGCCTCTAGCGTCTGCGTAAGTCCCGTTACGTTGCTTCGTATTCTCATGCATGGGAAAAAAACTTTCTGTGTGTGGGTGCTTCGTCCAATTCCTTTCCAAAACAAAAATAGGCGTCACTTTTCTG *************************************************************************************..................................((((...........)))).**************************************************************************************************************** |
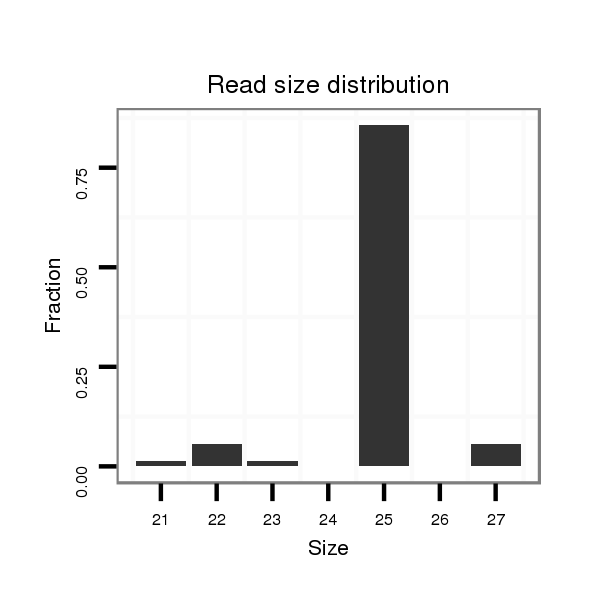
Read size | # Mismatch | Hit Count | Total Norm | Total | M044 female body |
V039 embryo |
V105 male body |
M058 embryo |
|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................TATCTACGCCACTGCCTCTAGCGTC................................................................................................................ | 25 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 |
| ......................................................................................................................TACCCCACCGCCTCTAGCGTC................................................................................................................ | 21 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| .........................................................................................................................................................................................AAACTTTCTGTGTGTGGGTGCTTCGTC....................................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................CGTATTCTCATGCATGGAAA.................................................................... | 20 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................TAGCGTCTGCGTAAGTCCCGTTACG.............................................................................................. | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 |
| ................................................................................................................................................................ACTCGTATTCTCATGCATG........................................................................ | 19 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 |
| .............................................................................................TATCCCCCCATATCAAAAAGCTATCTA................................................................................................................................... | 27 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................................CTGAGTCATATCCCCCCATATC................................................................................................................................................ | 22 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 |
| ...............................................................................ATTATTCTGAGTCATATCCCCCC..................................................................................................................................................... | 23 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .........................................................................................................CCAAAAAGCTATCTACGCCACTGC.......................................................................................................................... | 24 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| .................................................................................TATTCTGAGTCATATCCCCCC..................................................................................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
|
GTCATTATAATAATAATAATAATAACGTCTAACGCCTAATAATAGCGTCACATTTTATTTTGAATGTTTTTTTGTCCTATAATAAGACTCAGTATAGGGGGGTATAGTTTTTCGATAGATGCGGTGACGGAGATCGCAGACGCATTCAGGGCAATGCAACGAAGCATAAGAGTACGTACCCTTTTTTTGAAAGACACACACCCACGAAGCAGGTTAAGGAAAGGTTTTGTTTTTATCCGCAGTGAAAAGAC
****************************************************************************************************************..................................((((...........)))).************************************************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | V055 head |
M058 embryo |
M044 female body |
V039 embryo |
|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................................................................ACACCCACGAAGCCGGATA................................... | 19 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 |
| .........................................................................................................................................................................................................CCACGAAGGCAGTTAAGGAAA............................. | 21 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 |
| ..............................................................................................TAGGGGGGTATAGTTTTTCGAT....................................................................................................................................... | 22 | 0 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 |
| ................................................................................................................CGATATATAGGGTGACGGAGA...................................................................................................................... | 21 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................................CAGGGCAAGGGAACGAA........................................................................................ | 17 | 2 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 |
| ........................ACGTATAACGCCGAATAA................................................................................................................................................................................................................. | 18 | 2 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................TCCAGTCCAATGCAACGAA........................................................................................ | 19 | 3 | 19 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ..................................................................................................GGGGTATAGTTTTTCGATAGAT................................................................................................................................... | 22 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| ......................................................................TTTGTCCTATAATAAGACTCAGT.............................................................................................................................................................. | 23 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| ...................................................................................................GGGTATAGTTTTTCGATAGATGCGG............................................................................................................................... | 25 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| .................................GCCTAATAATAGCGTCACATTTTAT................................................................................................................................................................................................. | 25 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ....................................................................................................GGTATAGTTTTTCGATAGAGGCGGTG............................................................................................................................. | 26 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| .................................................................................................GGGGGTATAGTTTTTCGATAG..................................................................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| ...................................CTAATAATAGCGTCACATTTTATTTT.............................................................................................................................................................................................. | 26 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| .................................................................................................GGGGGTATAGTTTTTCGATAGGT................................................................................................................................... | 23 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ...................................................................................................GGGTATAGTTTTTCGATAGAT................................................................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droAna3 | scaffold_12916:14848127-14848377 - | dan_724 | CAGTAATATTATTATTATTATTATTGCAGATTGCGGATTATTATCGCAGTGTAAAATAAAACTTACAAAAAAACAGGATATTATTCTGAGTCATATCCC----------CCCATATCAAAAAGCTATCTACGCCACTGCCTCTAGCGTCTGCGTAAGTCCCGTTACGTTGCTTCGTATTCTCATGCATGGGAAAAAAACTTTCTGTGTGTGGGTGCTTCGTCCAATTCCTTTC----CAAAACAAAAATAGGCGTCACTTTTCTG |
| droBip1 | scf7180000396572:2151768-2151870 + | ----------------------AG-------------------------------------------------------------------------------------------------------------------------CGTCTGCGTAAGTTCCGTTAG---------------AATGCAAGCGTTAAAGCCTT----TGTGTGGGTGCTTCAGACCATTCTTTATTTAAATAAACAAAAATTGGCGTCAATTTTCTA | |
| droFic1 | scf7180000453563:3418-3548 + | ----------------------G------------GATTATTATCGCAGTGTAAAATATAACTTACAAAAAAACAGGATATTATTCTGAGTCATAACCCCCCCCCCCCCCCCATATCAAAAAGCTATCTACGCCACTGGCGCCAAAGCTGGGGAAAATGGCGCTA---------------------------------------------------------------------------------------------------- | |
| droEle1 | scf7180000487036:4098-4208 + | ----------------------ACTGTGCAACGCGGATTATTATCGCAGTGTAAAATAAAACTTACAAA-AAACAGGATATTATTCTGAGTCATATCCCC--------CCCCATATCAAAAAGCTATCTACGCCACTGTCTC--------------------------------------------------------------------------------------------------------------------------- | |
| droTak1 | scf7180000415308:4735-4779 - | ----------------------AT--------------------------------------------------------------------------------------------------------------------------------------------------------------------GGGAAAAAAATTATATCTTTGTTGGTTTTTATTACAATTCCTT---------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
Generated: 05/17/2015 at 11:02 PM