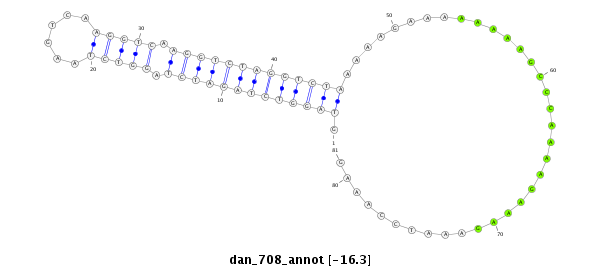
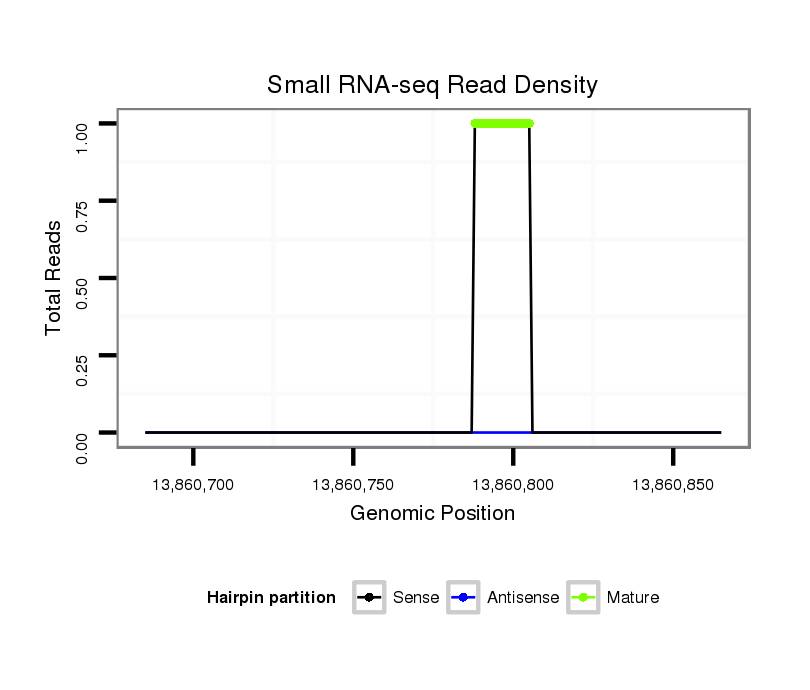
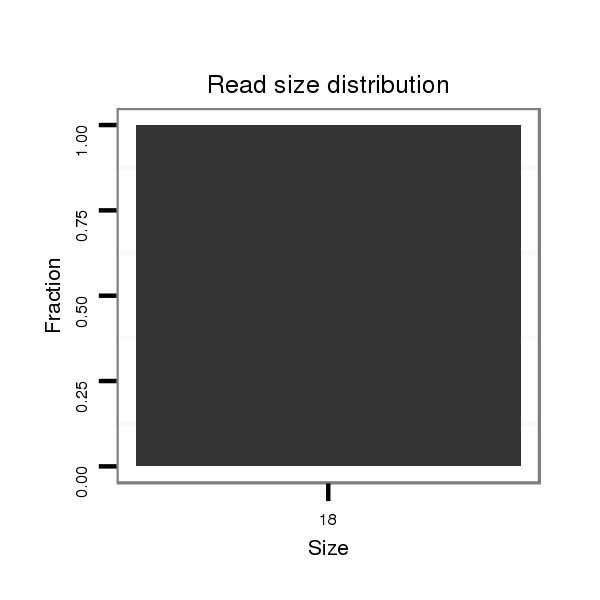
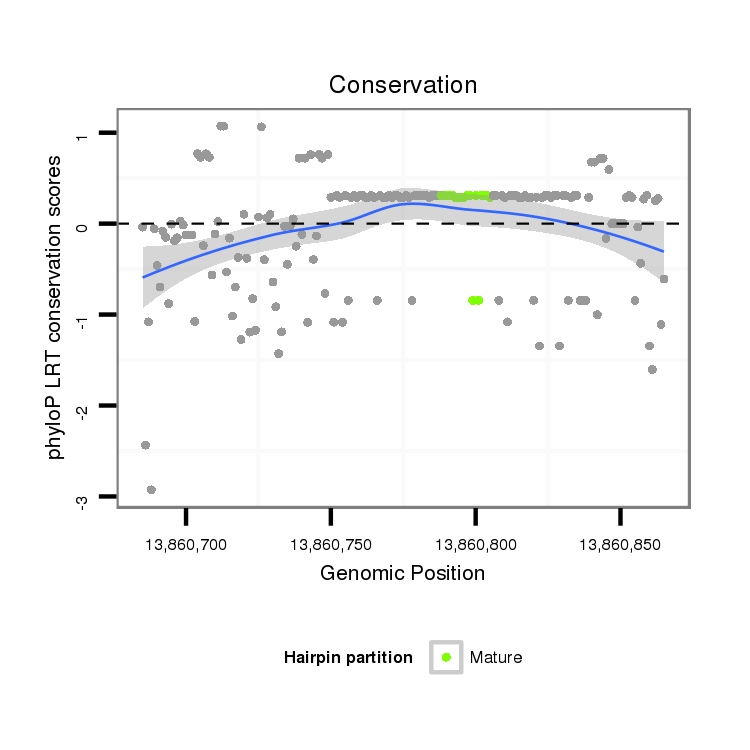
| .......................................................................................................AAAAAGCCCAAAAGAAAG............................................................ |
18 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
| .....AGATAAGATCCCGCACCAC............................................................................................................................................................. |
19 |
2 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
| ..................................................................................................AGAAAAAAAAACCCAAAAGAA.............................................................. |
21 |
1 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
| .................................................AATAGGTCTAGATCCAGG.................................................................................................................. |
18 |
2 |
2 |
0.50 |
1 |
0 |
0 |
1 |
0 |
| ...............................................................................................AGAAGAAAAAAAAGACCAAAAGA............................................................... |
23 |
2 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
| ...................................................................................................GAACAACCAGCCCAAAAGAAAGA........................................................... |
23 |
3 |
2 |
0.50 |
1 |
0 |
0 |
0 |
1 |
| ..............................................................................................AAAAAGAAGAGCAAGCCCAAAAGA............................................................... |
24 |
3 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
| ......................................................................................................AAAAAACCCCAAAAGAAAGA........................................................... |
20 |
1 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
| ................................................................................................................................................TAAGGATGTCGGGCAAGG................... |
18 |
2 |
4 |
0.25 |
1 |
0 |
1 |
0 |
0 |
| ...GATGATAAGATACCGACGCA.............................................................................................................................................................. |
20 |
3 |
4 |
0.25 |
1 |
0 |
1 |
0 |
0 |
| .....................................................................................................AAAAAAAGACCACAAGAAAGA........................................................... |
21 |
2 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
| ....................................................................................................AAAAAAAAGCGCAAAAGAA.............................................................. |
19 |
1 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
| ...................................................................................................GAAAAAAAAACCCAAAAGAA.............................................................. |
20 |
1 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
| ...............................................................................................AAAAGAAAAGCTAGCCCAAAAGA............................................................... |
23 |
3 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
| ...................................................................................................GAAAAAAAAGGCCCAAAGAAA............................................................. |
21 |
2 |
7 |
0.14 |
1 |
1 |
0 |
0 |
0 |
| ....................................................................................................AAAAAAAAGACCCAAAGAAGGAAA......................................................... |
24 |
3 |
10 |
0.10 |
1 |
1 |
0 |
0 |
0 |
| .............................................................................................TAAAAAGAGAAAGAAGCCCAC................................................................... |
21 |
3 |
11 |
0.09 |
1 |
1 |
0 |
0 |
0 |
| ......................................................................................................AAATAAACCCAAAAGAAAGAA.......................................................... |
21 |
2 |
11 |
0.09 |
1 |
1 |
0 |
0 |
0 |
| ....................................................................................................AAAAAAAACCCCAAAAGAA.............................................................. |
19 |
1 |
13 |
0.08 |
1 |
1 |
0 |
0 |
0 |
| .......................................................................................................AAAAAGCACTAAAGAAAGA........................................................... |
19 |
2 |
17 |
0.06 |
1 |
1 |
0 |
0 |
0 |
| ...............................................................................................AAAAGAAAAAAAAGCCGA.................................................................... |
18 |
1 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
| ....................................................................................................AAAAAAAAGCCGAAAAGAA.............................................................. |
19 |
1 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
| .................................................................................................AAGACAAAAAAGCCCAGAA................................................................. |
19 |
2 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
| ........................................................................................................AAAAGCCCAGAGGAAAGA........................................................... |
18 |
2 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
| ....................................................................................................AAAAAAAAACCCAAAAGAA.............................................................. |
19 |
1 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
| ...................................................................................GATCAAGGTGTAAAAAGAAA.............................................................................. |
20 |
3 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
| ........................................AAAGGGGAGAGTAGGTC............................................................................................................................ |
17 |
2 |
20 |
0.05 |
1 |
0 |
1 |
0 |
0 |
| ...................................................................................................GAAAAAAACACCCAAAAGAA.............................................................. |
20 |
2 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
| ............................................................................................CTAAAAAGAAAAAAAACAACAAAAGA............................................................... |
26 |
3 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
| ...............................................................................................................................AGAGGTCGTCGCGAGATG.................................... |
18 |
3 |
20 |
0.05 |
1 |
0 |
1 |
0 |
0 |