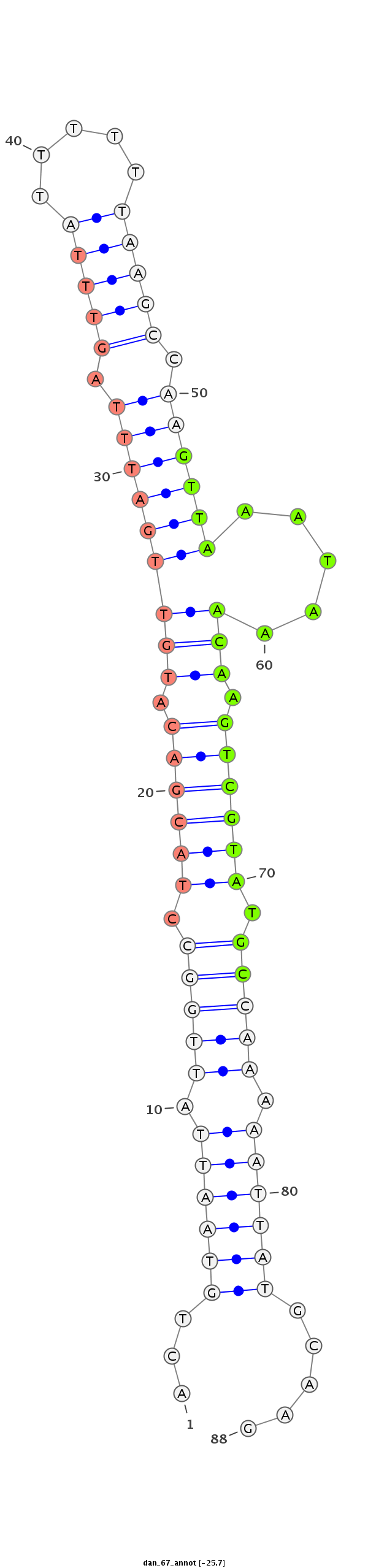



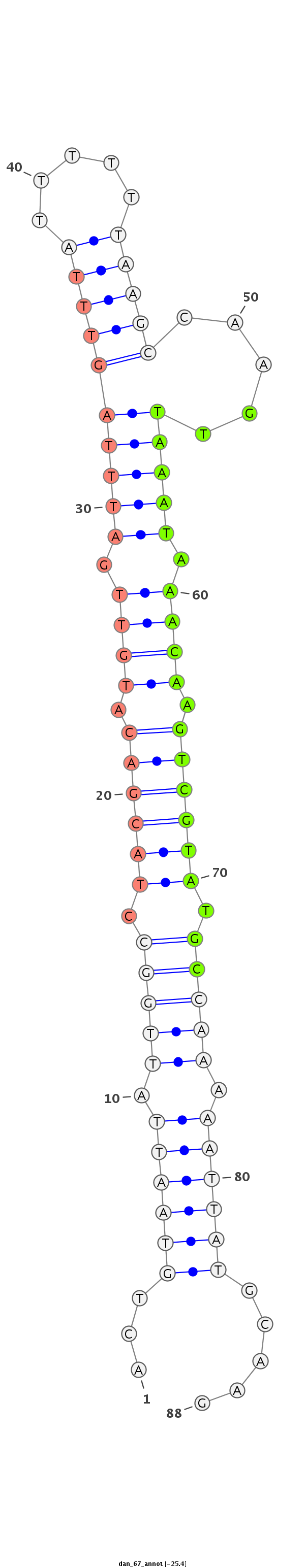
ID:dan_67 |
Coordinate:scaffold_13117:3425021-3425078 + |
Confidence:confident |
Class:Canonical miRNA |
Genomic Locale:antisense_to_intron |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -25.6 | -25.4 | -25.4 |
 |
 |
 |
Antisense to intron [Dana\GF20896-in]
No Repeatable elements found
|
GTGAAATAATTGTTAAATTAATTGGAGTTAAAAATACTGTAATTATTGGCCTACGACATGTTGATTTAGTTTATTTTTTAAGCCAAGTTAAATAAACAAGTCGTATGCCAAAAATTATGCAAGCCACAAGATCGATTCCATAAAAAAAAATATTTTTA
***********************************...((((((.(((((.((((((.(((((((((.(((((.....))))).)))))).....))).)))))).))))).)))))).....*********************************** |
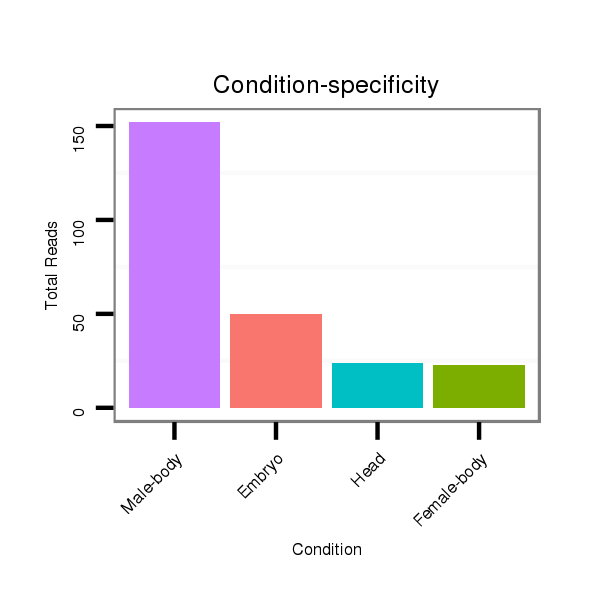
Read size | # Mismatch | Hit Count | Total Norm | Total | V105 male body |
M058 embryo |
M044 female body |
V055 head |
V106 head |
V039 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................GTTAAATAAACAAGTCGTATGC.................................................. | 22 | 0 | 1 | 104.00 | 104 | 47 | 36 | 3 | 6 | 7 | 5 |
| ......................................................................................GTTAAATAAACAAGTCGTATG................................................... | 21 | 0 | 1 | 97.00 | 97 | 89 | 2 | 2 | 3 | 1 | 0 |
| ..................................................CTACGACATGTTGATTTAGTTT...................................................................................... | 22 | 0 | 1 | 33.00 | 33 | 9 | 4 | 14 | 2 | 3 | 1 |
| ......................................................................................GGTAAATAAACAAGTCGTATGC.................................................. | 22 | 1 | 1 | 10.00 | 10 | 1 | 9 | 0 | 0 | 0 | 0 |
| ......................................................................................GTTAAATAAACAAGTCGTATGCT................................................. | 23 | 1 | 1 | 7.00 | 7 | 7 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................AGTTAAATAAACAAGTCGTATG................................................... | 22 | 0 | 1 | 4.00 | 4 | 3 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................GTTAAATAAACAAGTAGTATGC.................................................. | 22 | 1 | 1 | 3.00 | 3 | 1 | 0 | 0 | 2 | 0 | 0 |
| ..................................................CTACGACATGTTGATTTAGT........................................................................................ | 20 | 0 | 1 | 3.00 | 3 | 0 | 0 | 2 | 0 | 1 | 0 |
| ......................................................................................GTTAAATAAACAAGTCGTATGCA................................................. | 23 | 1 | 1 | 3.00 | 3 | 0 | 2 | 0 | 0 | 0 | 1 |
| ......................................................................................GTTAAATAAACAAGTCGTATTC.................................................. | 22 | 1 | 1 | 3.00 | 3 | 2 | 0 | 0 | 0 | 0 | 1 |
| .......................................................................................TTAAATAAACAAGTCGTATGC.................................................. | 21 | 0 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................TACGACATGTTGATTTAGTTT...................................................................................... | 21 | 0 | 1 | 2.00 | 2 | 1 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................GGTTAATAAACAAGTCGTATGC.................................................. | 22 | 2 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| ......................................................................................GTTAAATAAACAAGTCGTAT.................................................... | 20 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................GTTAAATAAACAAGTCGTATGT.................................................. | 22 | 1 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................GTTAGATAAACAAGTCGTATG................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................CTACGACAGGTTGATTTAGTTT...................................................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................GTTTAATAAACAAGTCATATGC.................................................. | 22 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................GTTAAATAAAAAAGTCGTATGC.................................................. | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .....................................................................................AGTTAAATAAACAAGTCGTATGT.................................................. | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................GTTAAATAAACAAGTCGCATGC.................................................. | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................GTTAAATAAACAAGTCGAATG................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................TTAAATAAACAAGTCGTATGCCT................................................ | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................TAATTAAACAAGTCGTATGC.................................................. | 20 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................GTTAAATAAACTAGTCGTATGC.................................................. | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................GCTAAATAAACAAGTAGTATGC.................................................. | 22 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................GTTAAATAAACACGTCGTAT.................................................... | 20 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................CTACGACATGTTGATTTAGTTTT..................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................GTTAAATAAACAAGTGGTATGC.................................................. | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................GGTAAATAAAAAAGTCGTATGCA................................................. | 23 | 3 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................TTAAATAAACAAGTCGTGTG................................................... | 20 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................GTTAAATAAACAAGTCGTA..................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................GTTAAATAAACAAGTCGAATGC.................................................. | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................GTTAAATAAACAAGTCGTATGCTA................................................ | 24 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................ATTTAAATAAACAAGTCGTATG................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................GTTAAATAAACAGGTCGTATGC.................................................. | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................................TTAAATAAACAAGTCGTATGCC................................................. | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................TGAAATAAACAAGTCGTATGC.................................................. | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................GTTAAATAAACAAGTCGTGTG................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................GTTAAATAAACAAGTCGTATGCAA................................................ | 24 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| .......................................................................................TGAAATAAACAAGTCGTATGCC................................................. | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................GTTAAATAAACAAGTCTTATGT.................................................. | 22 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................GTTAAATAAACAAGTCCTATG................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................TTTAATAAACAAGCCGTATGC.................................................. | 21 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..................................................CGACGACATGTTGATTTAGTTTAA.................................................................................... | 24 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................GTTAAATAAACAATTCGTATGC.................................................. | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................GTTAAATAGACAAGTCGTATGC.................................................. | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................GGCAAATAAACAAGTCGTATGC.................................................. | 22 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................GCTACGACGTGTTGATTTAGTTT...................................................................................... | 23 | 2 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................GTTAAATAAACGAGTCGTATG................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................CTACGACATGTTGATTTAGTT....................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................GATAAATAAACAAGTCGTATGC.................................................. | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................GTTAAATAAACAAGTCGTATT................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................GTCAAATAAACAAGTCGTATGCA................................................. | 23 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................GTTAAACAAACAAGTCGTATGC.................................................. | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................GGTAAATAAACAATTCGTATGC.................................................. | 22 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................GTTAAATAAACACGCCATATGC.................................................. | 22 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................GATAAATAAACAAGACGGATGC.................................................. | 22 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................TAATTGGAGATAAAAAGA.......................................................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................CAACGACATGTGGAATTAGTTT...................................................................................... | 22 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .......................................................................................................................................TTCCATACAAAAAAACATT.... | 19 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
|
CACTTTATTAACAATTTAATTAACCTCAATTTTTATGACATTAATAACCGGATGCTGTACAACTAAATCAAATAAAAAATTCGGTTCAATTTATTTGTTCAGCATACGGTTTTTAATACGTTCGGTGTTCTAGCTAAGGTATTTTTTTTTATAAAAAT
***********************************...((((((.(((((.((((((.(((((((((.(((((.....))))).)))))).....))).)))))).))))).)))))).....*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M044 female body |
M058 embryo |
|---|---|---|---|---|---|---|---|
| .................AATTAACCTCAATTTTGA........................................................................................................................... | 18 | 1 | 1 | 1.00 | 1 | 1 | 0 |
| ...................................................AGGCGGTATAACTAAATCAAA...................................................................................... | 21 | 3 | 8 | 0.13 | 1 | 0 | 1 |
| ..........................................................ACAACCTAATCAAATAAAAAA............................................................................... | 21 | 2 | 8 | 0.13 | 1 | 0 | 1 |
| ................GAATTAACCTCAATTTTGA........................................................................................................................... | 19 | 2 | 8 | 0.13 | 1 | 1 | 0 |
| .............ACCTAATTAACCTCAATTTTG............................................................................................................................ | 21 | 3 | 11 | 0.09 | 1 | 1 | 0 |
| ...............CGAATTAACCTCAATTTT............................................................................................................................. | 18 | 2 | 20 | 0.05 | 1 | 0 | 1 |
| Species | Coordinate | ID | Type | Alignment |
|---|---|---|---|---|
| droAna3 | scaffold_13117:3424971-3425128 + | dan_67 | confident | GTGAAATAATTGTTAAATTAA-TTGG-----------------------------------AGTTAAAAATA---------CTGTAATTATTGGCCTACGACATGTTGA-TTTAGTTTATTTTTTAAGCCAAGTTAAATAAACA--AGTCGTATGCCAAAAATTATGCAAGCCAC----------------------------AAGATCGATTC-CA--TA--------AAAAAAAATATT-----TTT-A |
| droBip1 | scf7180000395895:128435-128590 - | GAAATACTATT--------AA-TTGGAATCTTA----------------------------AGTCTAAAATA---------TTGCTGTGGTTGGCCTACGACATGTTGA-TTTAGTTTATGTCTGGAA-CAAATTAAATAAATA--AGTCGTTTGCCAAAAATTATGCAAA--------------------------------AACAACGATTT-AA--AA--------AAA-TAAATAATAAAAATAA-A | ||
| droEle1 | scf7180000491023:198326-198491 + | TGCAA-TAAATATATAATTTA-AAAG---------ATTCATTAAAGA-------------G--------------------------ATACAAGTCTTCGCTAAAAT---ATCAGTTTTTTTATTAACACAAA---------TATT------AGTCCAATAATGAAAT--GTCATACACCTTT--TAACTCGTTATACAATTTCAAACCGATTGACATTCA--------AACCAGAATATT-----TTTAT | ||
| droRho1 | scf7180000779985:163806-163966 - | ATAGG-TTTTTACTTACTTCC-TTAT------AAGATTAATATTGTATTTATATGTAACACCTACAAACATACAAAGAATT--------------TTTCAATATATTTTTTTCAGACTA---------------------CACC--AGACATAAGCCACA----------------AACATATGTAAATGGC-----------A---TTTGTTC-TT--TGTTATAAAATAATTTAA-----------T-A | ||
| droYak3 | X:6797531-6797649 + | ACGAAGGAATT--------AAATCG-----------------------------------GATACGATCAAACTGGAAATTTAGATTTT---------------------------------------TCAAGCAAAAGAAACATTATTTTTAATCTAAAAATAAACTTAGCCAGGAATCTTT--AAATTG----------------------------TA--------TAAATAAA-----------T-C |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
Generated: 10/20/2015 at 06:47 PM