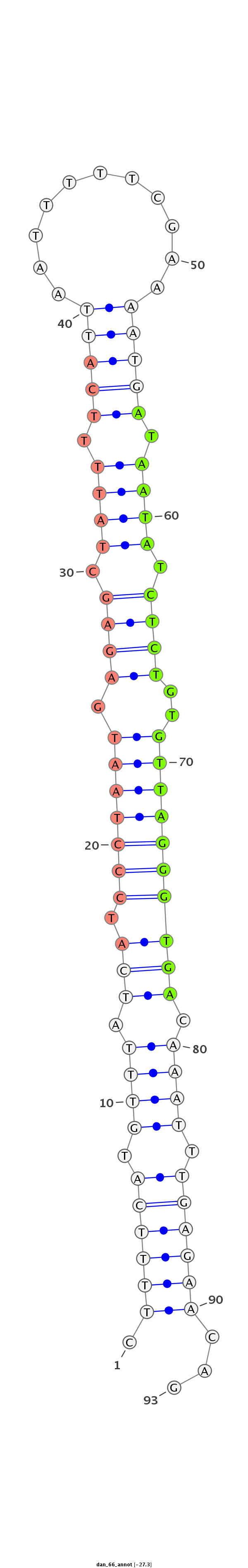
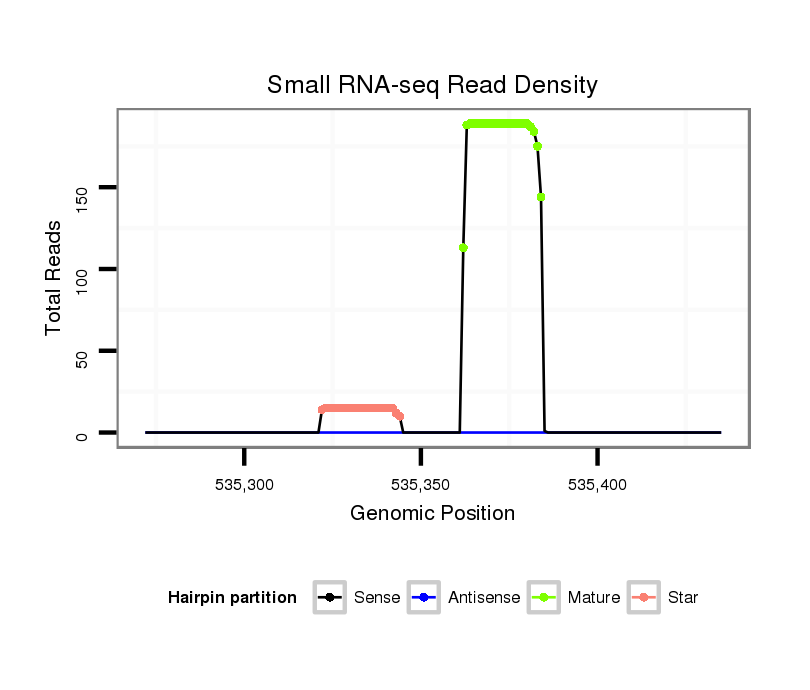
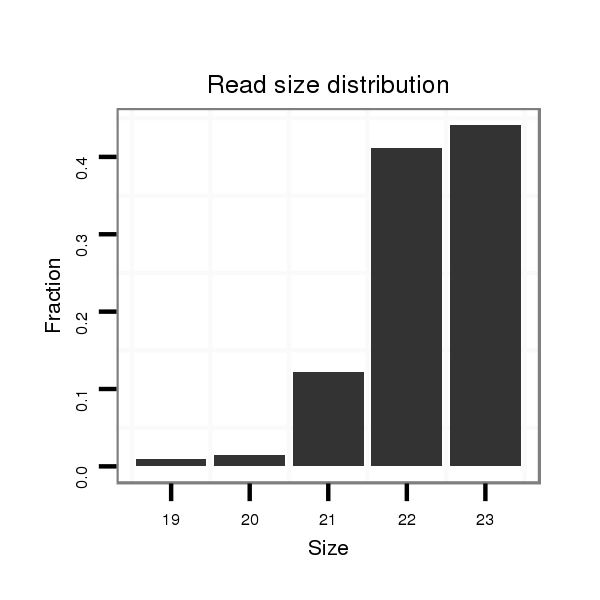
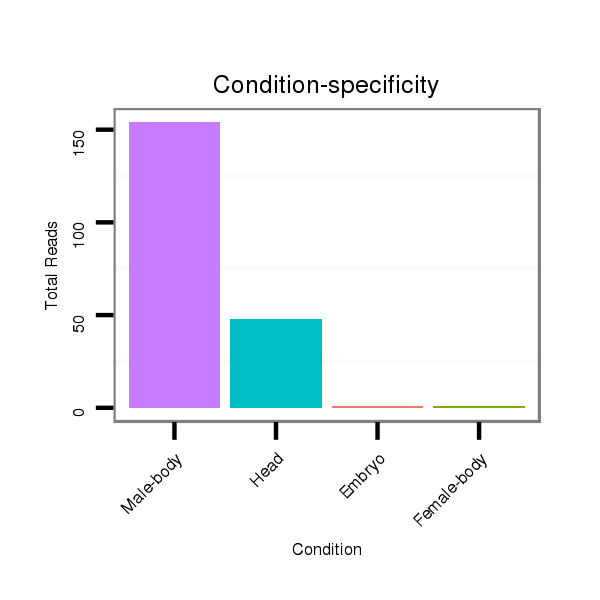
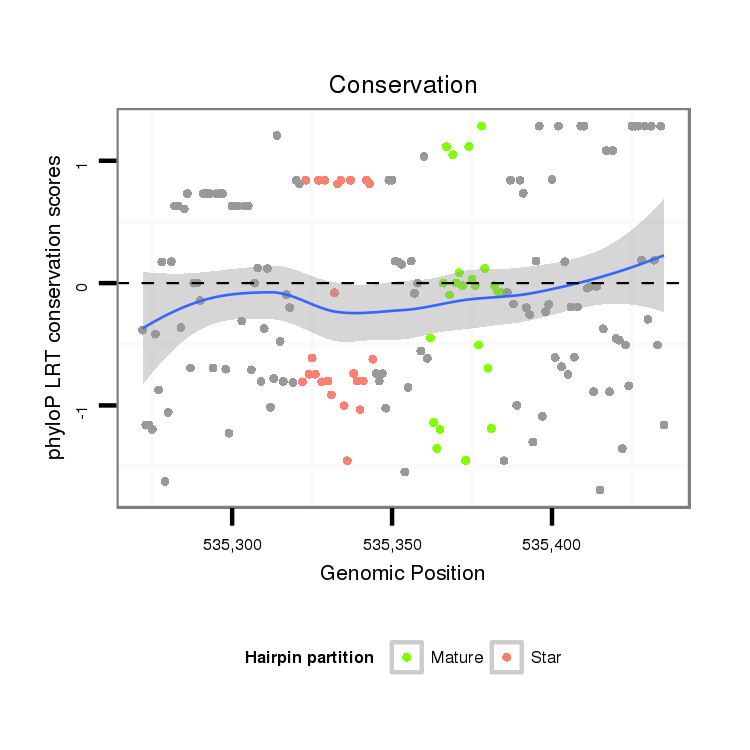
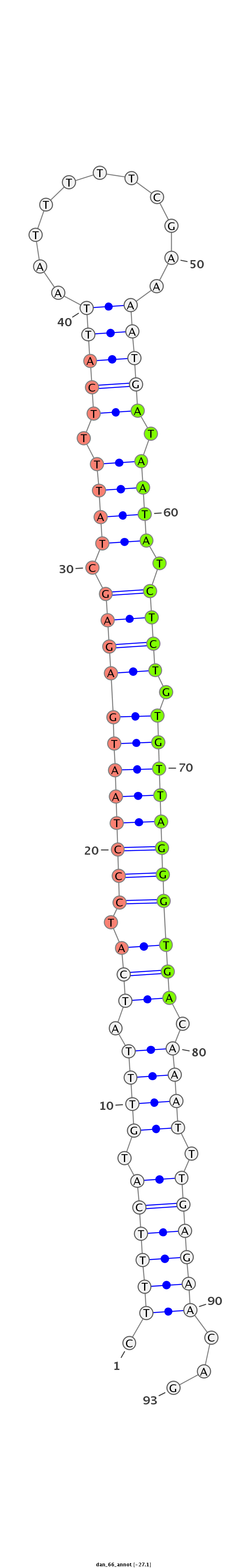
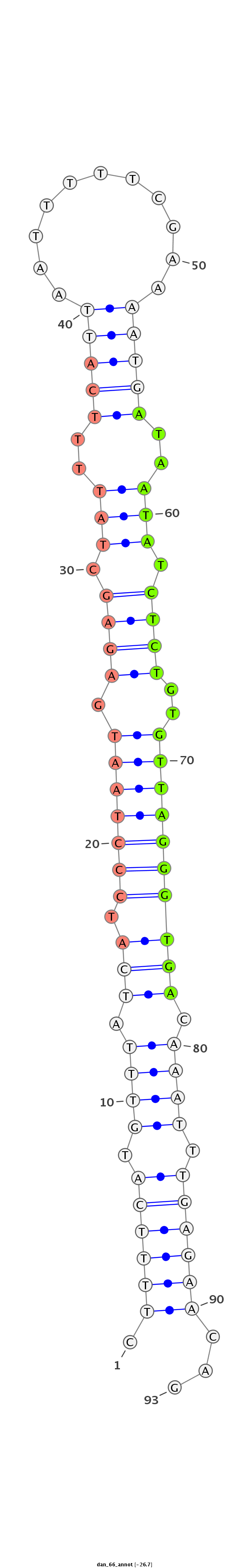
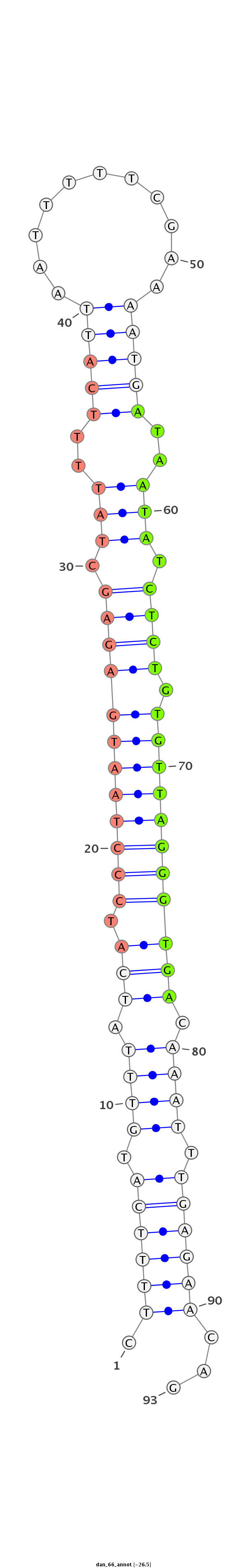
ID:dan_66 |
Coordinate:scaffold_12903:535322-535385 + |
Confidence:confident |
Class:Testes-restricted |
Genomic Locale:intergenic |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -27.1 | -26.7 | -26.5 |
 |
 |
 |
intergenic
No Repeatable elements found
|
TTTTTTATTGAATCTTTTATTTTTTAGTTATCTTCCTTTTCATGTTTATCATCCCTAATGAGAGCTATTTTCATTAATTTTTCGAAAATGATAATATCTCTGTGTTAGGGTGACAAATTTGAGAACAGTCTACAAAATAAAGTGGTGTTTATATATATTTTTAT
***********************************.((((((.((((.(((.(((((((.((((.((((.(((((...........))))).)))).))))..)))))))))).)))).))))))...************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | V105 male body |
V055 head |
V106 head |
M044 female body |
M058 embryo |
V039 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ..........................................................................................ATAATATCTCTGTGTTAGGGTGA................................................... | 23 | 0 | 1 | 80.00 | 80 | 64 | 12 | 3 | 1 | 0 | 0 |
| ...........................................................................................TAATATCTCTGTGTTAGGGTGA................................................... | 22 | 0 | 1 | 62.00 | 62 | 53 | 6 | 3 | 0 | 0 | 0 |
| ..........................................................................................ATAATATCTCTGTGTTAGGGTG.................................................... | 22 | 0 | 1 | 19.00 | 19 | 11 | 0 | 8 | 0 | 0 | 0 |
| ...........................................................................................TAATATCTCTGTGTTAGGGTG.................................................... | 21 | 0 | 1 | 12.00 | 12 | 9 | 2 | 1 | 0 | 0 | 0 |
| ..................................................ATCCCTAATGAGAGCTATTTTCA........................................................................................... | 23 | 0 | 1 | 9.00 | 9 | 2 | 0 | 7 | 0 | 0 | 0 |
| ..........................................................................................ATAATATCTCTGTGTTAGGGT..................................................... | 21 | 0 | 1 | 9.00 | 9 | 6 | 2 | 1 | 0 | 0 | 0 |
| ...........................................................................................TAATATCTCTGTGTTAGGGTGT................................................... | 22 | 1 | 1 | 5.00 | 5 | 5 | 0 | 0 | 0 | 0 | 0 |
| ..................................................ATCCCTAATGAGAGCTATTTT............................................................................................. | 21 | 0 | 1 | 3.00 | 3 | 2 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................ATAATATCTCTGTGTTAGGGTGAA.................................................. | 24 | 1 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................ATAATATCTCTGTGTTAGGG...................................................... | 20 | 0 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| ..................................................ATCCCTAATGAGAGCTATTTTC............................................................................................ | 22 | 0 | 1 | 2.00 | 2 | 0 | 1 | 0 | 0 | 1 | 0 |
| ..........................................................................................ATAATATCTCTGTGTTAGG....................................................... | 19 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................CAATATCTCTGTGTTAGGGTGA................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................ATAATATCTCTGTGTTAGGGAGA................................................... | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................TTATATCTCTGTGTTAGGGTG.................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................TAATATCTCTGTTTTAGGGTGA................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................TAATATCTCTCTGTTAGGGTGA................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................ATAATATCTCTTTGTTAGGGTGA................................................... | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................TAATATCTCTGTTTTAGGGTG.................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................TAATGTCTCTGTGTTAGGGTGA................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................ATAATATCTCTGTGTTAGGGGGA................................................... | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................ATAATATCTCTGTGTTAGTGT..................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................ATAATATCTCTGTGTTATGGTGA................................................... | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................ATACCTACTGAGAGCTATTTT............................................................................................. | 21 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................TAATATCTCTGTGTTAGGGTGAC.................................................. | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................TAATATCTCTGTGTTAGGGTCA................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................TAATATCTCTGTGTTAGGTTGA................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................AATATCTCTGTGTTAGGGTGA................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................TCCCTAATGAGAGCTATTTTCA........................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................ATAATATCTCTCTGTTAGGGTGA................................................... | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................ATAATATCGCTGTGTTAGGGG..................................................... | 21 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................ATAATATCTCTGTGCTAGGGTGA................................................... | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................GTAATATCTCTGTGTTAGGGTGA................................................... | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................ATAATATCTCTGTGTTAGGGTGG................................................... | 23 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................TAATATCACTGTGTTAGGGTGA................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................ATCCCTAATGAGAGCTATTTTCT........................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................ATAATATCTCTGTGTTAGGGTGTTA................................................. | 25 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................TAATATCTCTGTGATAGGGTG.................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................TAATATCTTTGTGTTAGGGT..................................................... | 20 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...........................................................................................TAATATCTCTGTGTTAGGGTGG................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................ATAACCTCTGTGTTAGGGTGA................................................... | 21 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................ATAATATCTCTGCGTTAGGGTGC................................................... | 23 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................TAATATCTCTGTGTTAAGGTGA................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................TAATATCTCTGTGTTAGGGTGAT.................................................. | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................ATAATATCTCTGTGTTAGGGTGT................................................... | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................TAATATCTCTGTGGTAGGGTGA................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................ATAATATCTCTGTGTCAGGGTGA................................................... | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................TAATATCTCTGTGTTAGGATGA................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................TAATACGTCTGTGTTAGTGTGA................................................... | 22 | 3 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................GAACAGGCTACAAAAGAA........................ | 18 | 2 | 18 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
|
AAAAAATAACTTAGAAAATAAAAAATCAATAGAAGGAAAAGTACAAATAGTAGGGATTACTCTCGATAAAAGTAATTAAAAAGCTTTTACTATTATAGAGACACAATCCCACTGTTTAAACTCTTGTCAGATGTTTTATTTCACCACAAATATATATAAAAATA
************************************.((((((.((((.(((.(((((((.((((.((((.(((((...........))))).)))).))))..)))))))))).)))).))))))...*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V105 male body |
V039 embryo |
M058 embryo |
V055 head |
|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................CACCACAACGATATATCAAAA.. | 21 | 3 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 |
| ................................AAGGGAAAGTACAAAGAGTA................................................................................................................ | 20 | 2 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 |
| ....................AAAAATGAAGAGAAGGAAAA............................................................................................................................ | 20 | 2 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| ...............................................................CGATAAAAGTATTAACAAAGC................................................................................ | 21 | 3 | 17 | 0.06 | 1 | 0 | 0 | 1 | 0 |
| ............................................................................................TAATAGAGCCACAATACCA..................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Type | Alignment |
|---|---|---|---|---|
| droAna3 | scaffold_12903:535272-535435 + | dan_66 | confident | TTTTTTATTGAATCTTTTATTTTTTAGTTATCTTCCTTTTCATGTTTATCATCCCTAATGAGAGCTATTTTCATTAATTTTTCGAA-----------AATGATAATATCTCTGTGT---TAGGGTGACAAATTTGAGAACAGT------------CTACAAAATAAAGTGGTGT-TTATATATATTTTTAT |
| droBip1 | scf7180000395952:1943-2080 - | TAACTAAAAA----TC--ATTTCTTAAG-----------------TTTTCTTTTTTTAATAGAAGTGAGATCGCAGTTTCTTTCAA-----------A----GGCTATCTCACTGAAATTAGAAAGAGAAACT---TGAGA------------ACCTGTCAAATAATAACATATCAACTGTATTTTTATAA | ||
| droKik1 | scf7180000302798:211533-211619 - | GGGAAAGGTA--------------------------TGGTGTTTGAGA-------------------------------TCGGGTGAGTTATATTC-----------------------GATTT------------AAATAAT------------TTGTAAGATATAATAATATTTAAGATATATCTTGAG | ||
| droTak1 | scf7180000415843:977999-978130 - | TTTTCTACGAAAACTT--GTTTTTTAAATATTTTG-AAGCACTTAAGT-------------------------------TTTAGAGAATTATATTTATGTTCTCGAAGCACTTTAT---AATTAT------------AATTGTTTGAATAACGCTGTATATTTTA----------TATATTATATTTTAAT | ||
| droGri2 | scaffold_14822:226493-226630 - | ATTATTAGTA--------------------------TTTTCATATTTATCTTTCTTTATGTGATTTATTTTCGTTACTTTTTTTAA-----------ATTAAATGTATCTTTCTGT---TATTGTATTTACATTATTAATG------------GTTTATTTTTTAAATTTATTT-ATTTATATATATTTAT |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 10/20/2015 at 06:45 PM