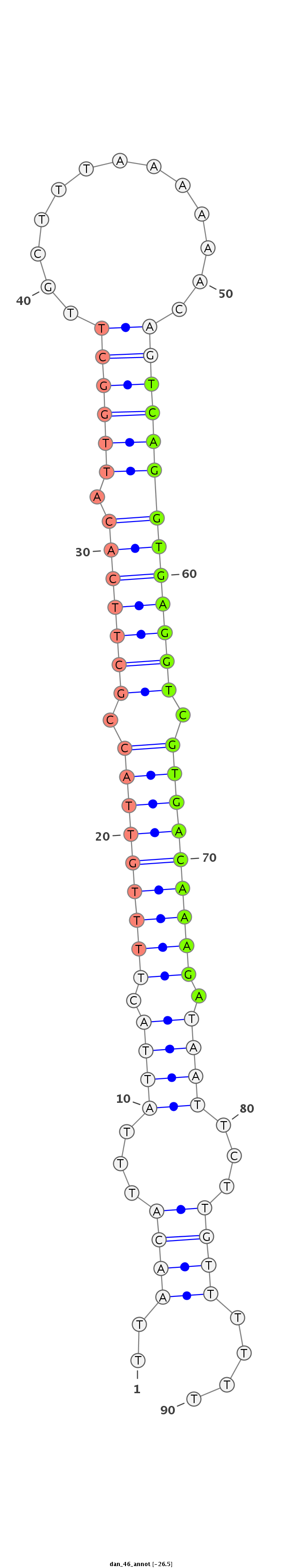
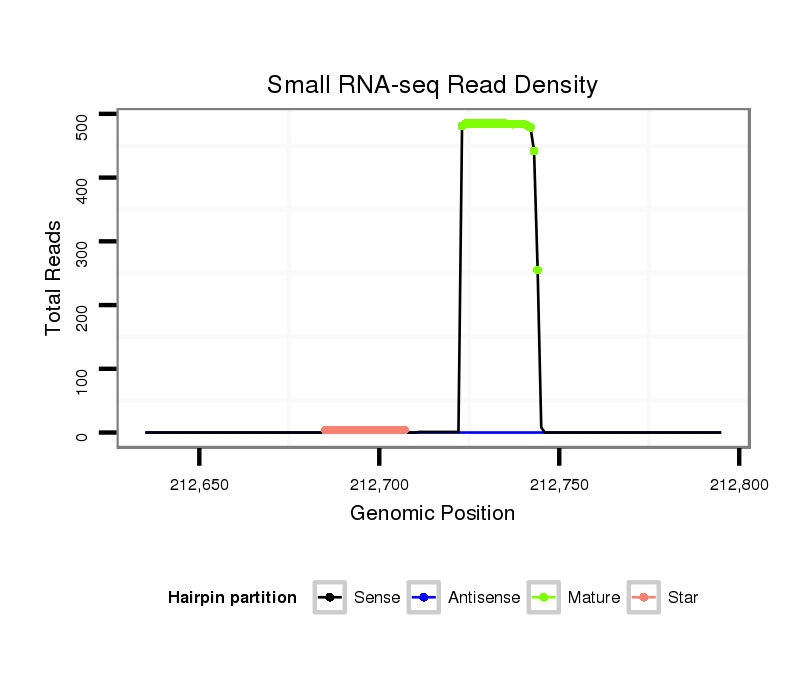
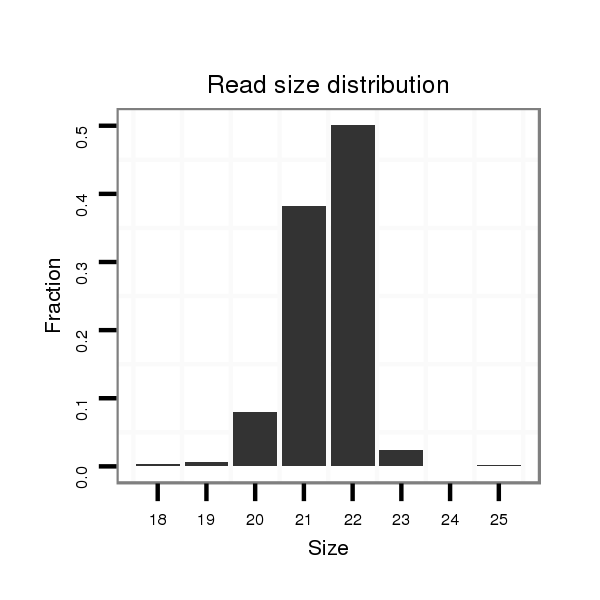
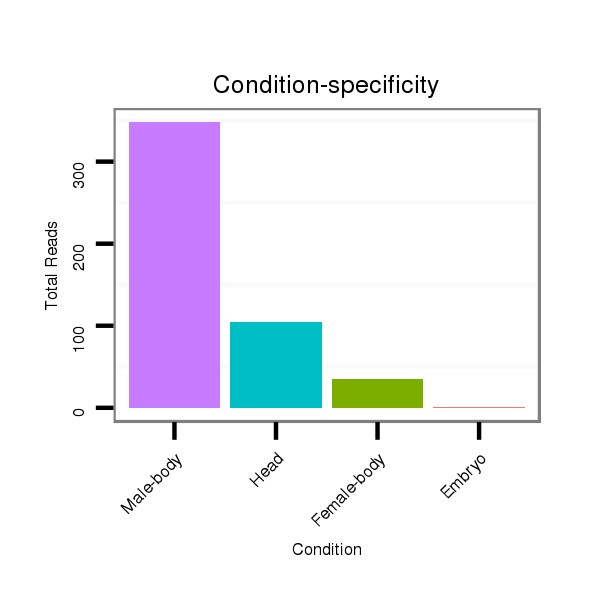
| ........................................................................................TCAGGTGAGGTCGTGACAAAGA................................................... |
22 |
0 |
1 |
245.00 |
245 |
179 |
46 |
14 |
6 |
0 |
| ........................................................................................TCAGGTGAGGTCGTGACAAAG.................................................... |
21 |
0 |
1 |
185.00 |
185 |
132 |
34 |
11 |
7 |
1 |
| ........................................................................................TCAGGTGAGGTCGTGACAAA..................................................... |
20 |
0 |
1 |
37.00 |
37 |
27 |
7 |
2 |
1 |
0 |
| ........................................................................................TCAGGTGAGGTCGTGACAAAGAA.................................................. |
23 |
1 |
1 |
15.00 |
15 |
11 |
1 |
1 |
2 |
0 |
| ........................................................................................TCAGGTGAGGTCGTGACAAAGAT.................................................. |
23 |
0 |
1 |
8.00 |
8 |
4 |
2 |
1 |
1 |
0 |
| ........................................................................................TCAGGTGAGGTCGTGACAAAGT................................................... |
22 |
1 |
1 |
4.00 |
4 |
3 |
1 |
0 |
0 |
0 |
| ..................................................TTTGTTACCGCTTCACATTGGCT........................................................................................ |
23 |
0 |
1 |
4.00 |
4 |
1 |
0 |
3 |
0 |
0 |
| ........................................................................................TCAGGTGAGGTCGTGACAAAT.................................................... |
21 |
1 |
1 |
3.00 |
3 |
1 |
1 |
0 |
1 |
0 |
| ........................................................................................TCAGGTGAGGTCGTGACAAAGAAA................................................. |
24 |
1 |
1 |
3.00 |
3 |
2 |
1 |
0 |
0 |
0 |
| ........................................................................................TCAGGTGAGGTCGTGACAA...................................................... |
19 |
0 |
1 |
3.00 |
3 |
3 |
0 |
0 |
0 |
0 |
| ........................................................................................TCAGGTGAGGTCGTGACAAATA................................................... |
22 |
1 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
| ........................................................................................TCAGGTGAGGTCGTGACAAAA.................................................... |
21 |
1 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
| ........................................................................................TCAGGTGAGGTCGTTACAAAG.................................................... |
21 |
1 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
| ........................................................................................TCAGGTGAGGTCGTGACA....................................................... |
18 |
0 |
1 |
2.00 |
2 |
0 |
1 |
1 |
0 |
0 |
| ........................................................................................TCAGGTGAGGTGGTGACAAAGA................................................... |
22 |
1 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
| .........................................................................................CAGGTGAGGTCGTGACAAAG.................................................... |
20 |
0 |
1 |
2.00 |
2 |
0 |
0 |
2 |
0 |
0 |
| .........................................................................................CAGGTGAGGTCGTGACAAAGA................................................... |
21 |
0 |
1 |
2.00 |
2 |
1 |
0 |
1 |
0 |
0 |
| ........................................................................................TCAGGTGAGGTAGTGACAAAGA................................................... |
22 |
1 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
| ........................................................................................TCAGGTGAGGTTGTGACAAAGA................................................... |
22 |
1 |
1 |
2.00 |
2 |
2 |
0 |
0 |
0 |
0 |
| ........................................................................................TCAGGTGAGGTCTTGACA....................................................... |
18 |
1 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
| ........................................................................................TCGGGTGAGGTCGTGACAAAG.................................................... |
21 |
1 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
| ........................................................................................TCAGGTGAGGTCGTGACAAT..................................................... |
20 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................................................................................TCAGGTGAGGTCTTGACAAAGA................................................... |
22 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................................................................................TCAGGTGAGGTAGTGACAAAC.................................................... |
21 |
2 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
| ........................................................................................TCAGGTCAGGTCGTGACAAAT.................................................... |
21 |
2 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................................................................................TCAGGTGAGGTCGTGACAAAAA................................................... |
22 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................................................................................TCAGGTGAGGTCGTTACAAAGA................................................... |
22 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ............................................................................TTTAAAAAACAGTCAGGTGAGGTCG............................................................ |
25 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................................................................................TCAGGTGAGGTCGTGACAAAAAT.................................................. |
23 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................................................................................TCAGGTGAGGTCGTGACAAAGAAT................................................. |
24 |
2 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................................................................................TCAGGTCAGGTCGTGACAAAGA................................................... |
22 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................................................................................TCATGTGAGGTCGTTACAAAGA................................................... |
22 |
2 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................................................................................TCAGGTGAGGTCGTGACAACGA................................................... |
22 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................................................................................TCAGGTGAGGTCGTGACAAAC.................................................... |
21 |
1 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
| ........................................................................................TCAGGTGAGGTCGTGACGAAGAA.................................................. |
23 |
2 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
| ........................................................................................TCAGGTGAGCTCGTGACAAAG.................................................... |
21 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................................................................................TCAGGTGGGGTCGTGACAAAG.................................................... |
21 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................................................................................TCAGGTGAGGGCGTGACAAAG.................................................... |
21 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................................................................................TCAGGTGAGGTCGGGACAAAG.................................................... |
21 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................................................................................TCAGGTGAGGCCGTGACAAAGA................................................... |
22 |
1 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
| ........................................................................................TCAGGTGAGGTCGTGACAAGGAAAA................................................ |
25 |
2 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................................................................................TCAGGTGACGTCGAGACAAAGA................................................... |
22 |
2 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .........................................................................TGCTTTGAAAAGCAGTCTGGTG.................................................................. |
22 |
3 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
| ........................................................................................TCAGGTGAGGTAGTGACAAAGAAA................................................. |
24 |
2 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................................................................................TCAGGTGAGGACGTGACAAAG.................................................... |
21 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ................................................................................................................................CTCTTTAAACCACTAAGTTTA............ |
21 |
2 |
1 |
1.00 |
1 |
0 |
0 |
0 |
0 |
1 |
| ........................................................................................TCAGGTGAGGTCGTGACTAAG.................................................... |
21 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................................................................................TCAGGTGATGTCGTGACAAAGA................................................... |
22 |
1 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
| ........................................................................................TCAGGTGAGGTCCTGACAAAGA................................................... |
22 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................................................................................TCAGGTGAAGTCGTGACAAAG.................................................... |
21 |
1 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
| ........................................................................................TCAGGTGAGGTCGAGACGAAGA................................................... |
22 |
2 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................................................................................TCAGGTGACGTCGTGACAAAGA................................................... |
22 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................................................................................TCAGGTGAGGTCGTGACAAAGAAC................................................. |
24 |
2 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................................................................................TCAGGTGAGGTCATGACAAAG.................................................... |
21 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ................................................................................AAATACCGTCAGGTGAGGTGGT........................................................... |
22 |
3 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
| ........................................................................................TCAGGTCAGGTCGTGACAAAG.................................................... |
21 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................................................................................TCAGGTGAGGTCGTGCCAAAGA................................................... |
22 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................................................................................TCAGGTGAGGTCGTGATAAAG.................................................... |
21 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................................................................................TCAGGTGAGGACGTGACAAAGA................................................... |
22 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................................................................................TAAGGTGAGGTCGTGACAAAG.................................................... |
21 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................................................................................CCAGGTGAGGTCGTGACAAAG.................................................... |
21 |
1 |
1 |
1.00 |
1 |
0 |
0 |
0 |
1 |
0 |
| ..................................................................................AAACAGGCAGGTGAGGTGCTG.......................................................... |
21 |
3 |
2 |
0.50 |
1 |
0 |
0 |
1 |
0 |
0 |
| ........................................................................................TCAGGTGAGGTCGCGACA....................................................... |
18 |
1 |
2 |
0.50 |
1 |
1 |
0 |
0 |
0 |
0 |
| ......................................................................................AGTCAGGGGAGGTAGTGACGA...................................................... |
21 |
3 |
2 |
0.50 |
1 |
0 |
0 |
0 |
1 |
0 |
| .....AAAGATAAATGCAAAAGATGAAG..................................................................................................................................... |
23 |
3 |
3 |
0.33 |
1 |
0 |
0 |
0 |
1 |
0 |
| ......................................................................................AGCCAGGTGAGGTAGTGACGA...................................................... |
21 |
3 |
5 |
0.20 |
1 |
0 |
0 |
0 |
1 |
0 |
| ...........................................................................................GGTGAAGTCGTGACAAGGT................................................... |
19 |
3 |
11 |
0.18 |
2 |
2 |
0 |
0 |
0 |
0 |
| ..................................................................................AAACAGGCAGGTGAGGTGCT........................................................... |
20 |
3 |
6 |
0.17 |
1 |
0 |
0 |
1 |
0 |
0 |
| ..........................................................................................AGGGGAGGTCGTGACGA...................................................... |
17 |
2 |
15 |
0.13 |
2 |
0 |
0 |
0 |
2 |
0 |
| ......................................................................................................GACAAAGATAAATCTTTTT........................................ |
19 |
2 |
13 |
0.08 |
1 |
0 |
0 |
1 |
0 |
0 |
| ................................................................................AAATACAGTCAGGTGAAG............................................................... |
18 |
2 |
16 |
0.06 |
1 |
0 |
0 |
0 |
1 |
0 |
| ............................................................CTTCACATTGATTTGCTTTAT................................................................................ |
21 |
3 |
16 |
0.06 |
1 |
0 |
0 |
1 |
0 |
0 |
| ..........................................................................................AGGGGAGCTCGTGACAA...................................................... |
17 |
2 |
16 |
0.06 |
1 |
0 |
0 |
0 |
1 |
0 |
| ..................................................................................AAGCAGCAAGGTGAGGTCG............................................................ |
19 |
3 |
20 |
0.05 |
1 |
0 |
0 |
0 |
1 |
0 |