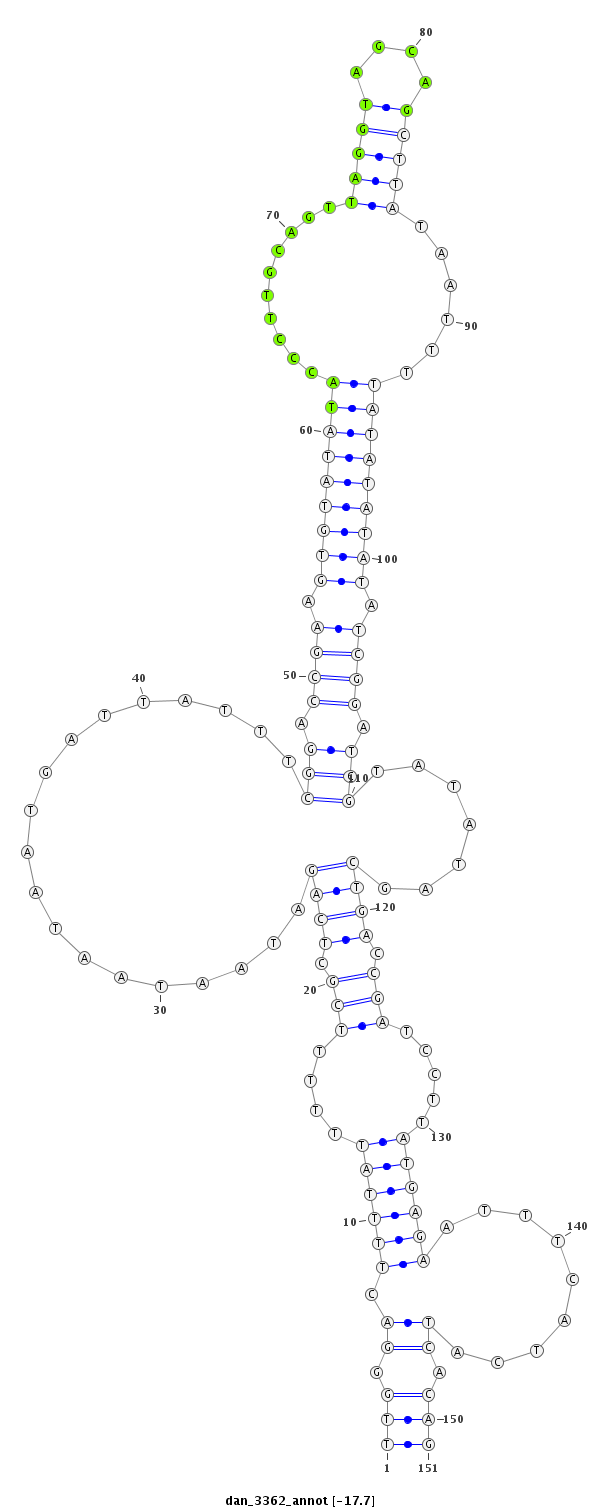
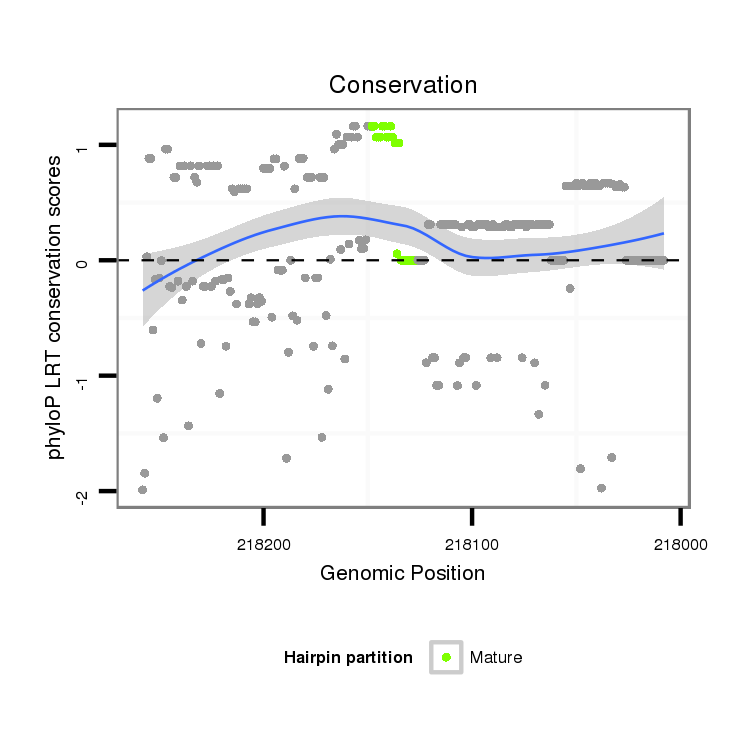
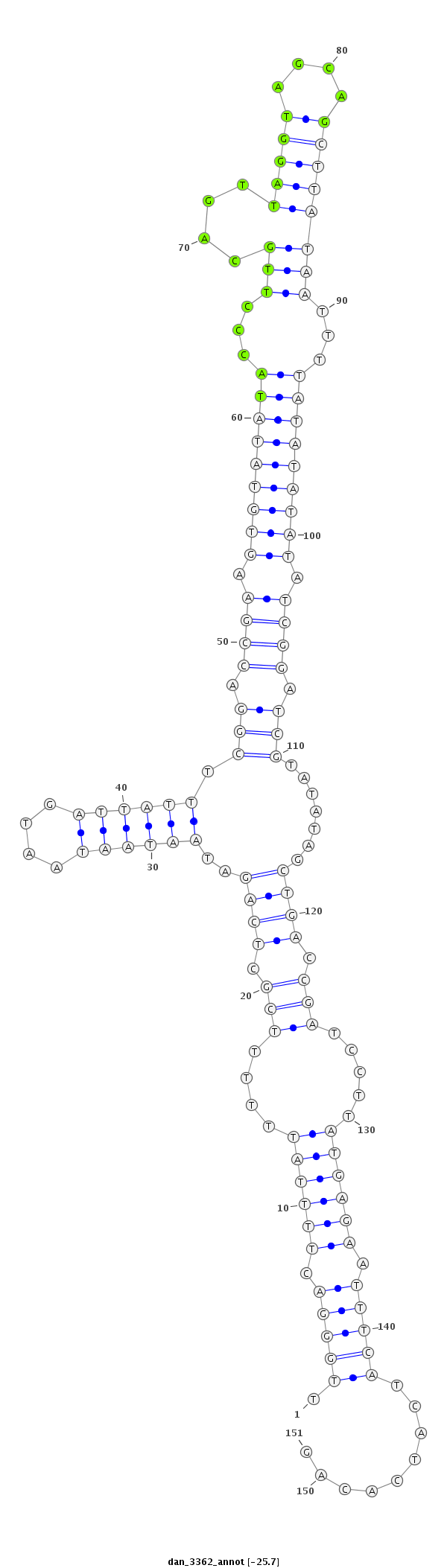
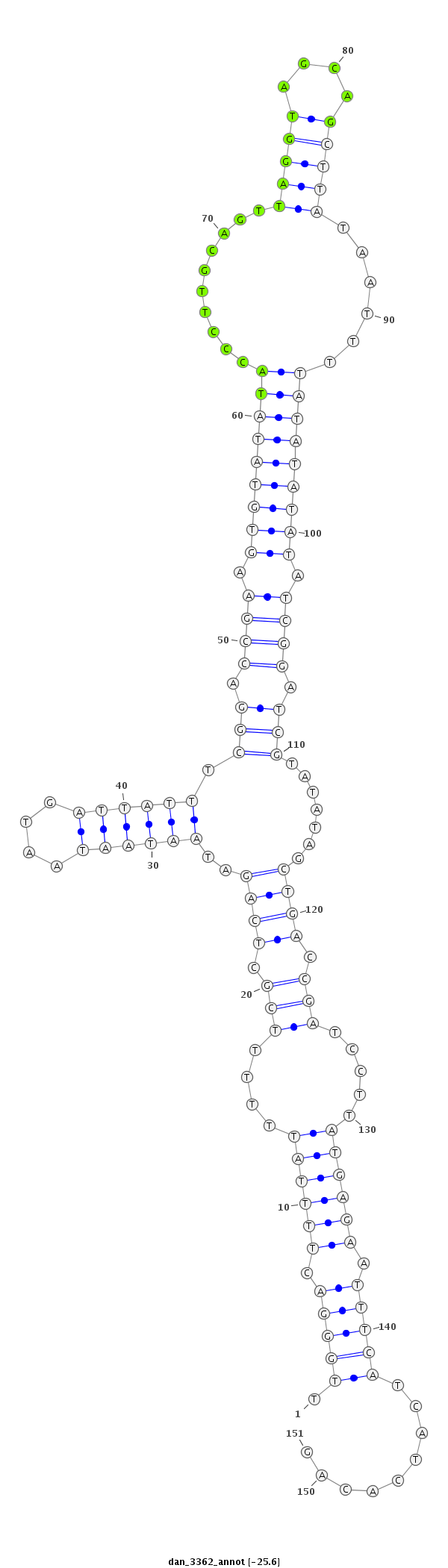
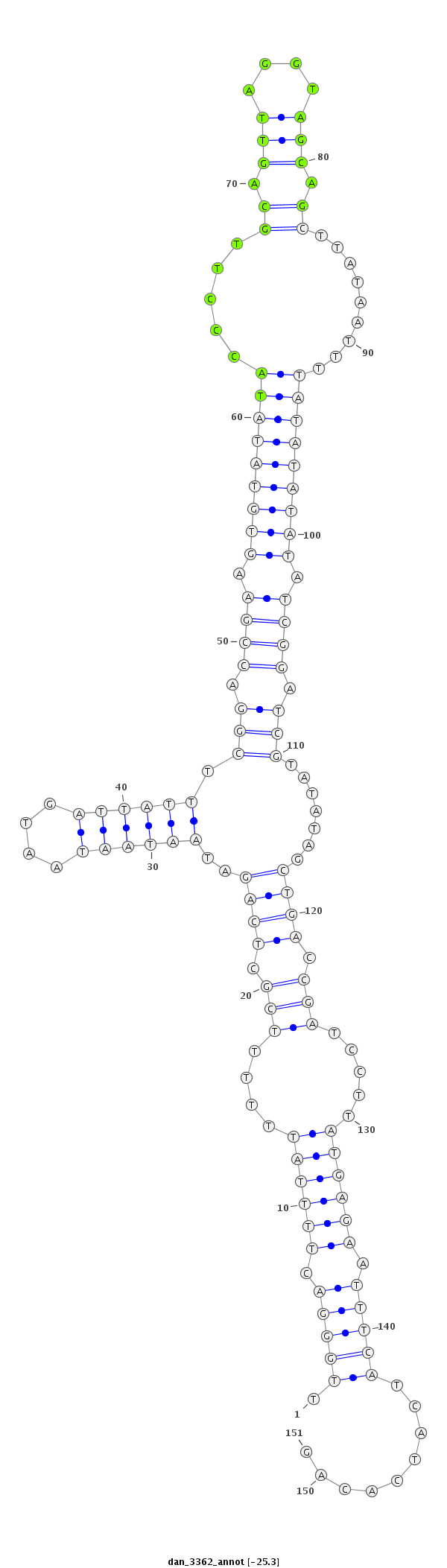
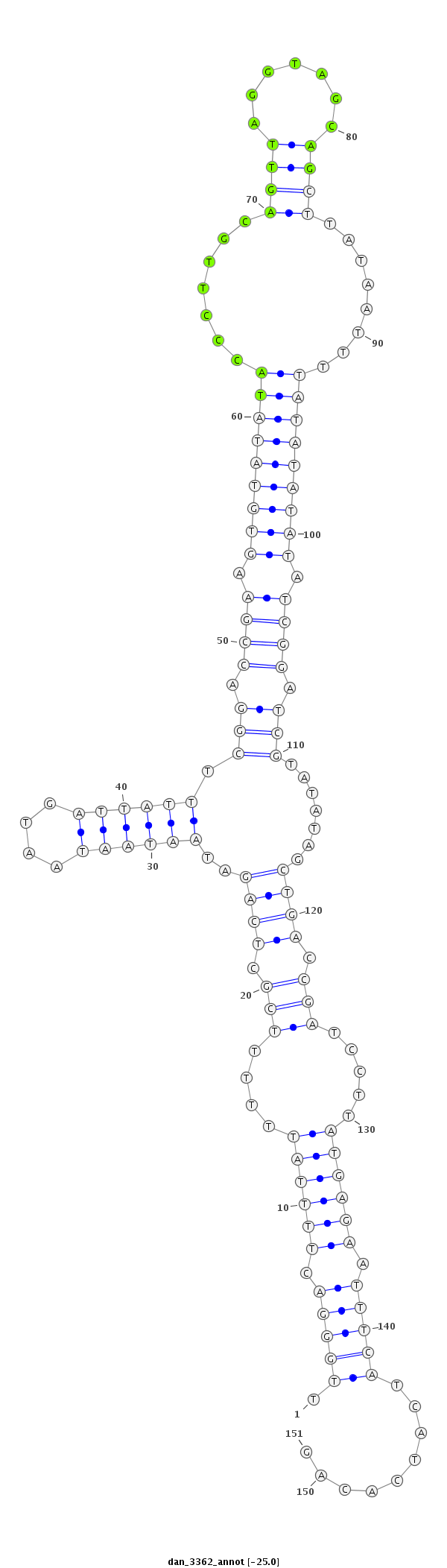
ID:dan_3362 |
Coordinate:scaffold_13499:218058-218208 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -25.7 | -25.6 | -25.3 | -25.0 |
 |
 |
 |
 |
CDS [Dana\GF23829-cds]; exon [dana_GLEANR_86:2]; intron [Dana\GF23829-in]
| Name | Class | Family | Strand |
| Hoana8 | DNA | hAT-hobo | + |
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## ACATAGCCCGCAATCATGAATTAAGTGATATTTTTGACCATAATGATTATTTGGGACTTTTATTTTTTCGCTCAGATAATAATAATGATTATTTCGGACCGAAGTGTATATACCCTTGCAGTTAGGTAGCAGCTTATAATTTTATATATATATCGGATCGTATATAGCTGACCGATCCTTATGAGAATTTCATCATCACAGGTGCCGGTCATCGCACAGGTTTTTATGCACGAAACTTCGCAGTTCGCGAC **************************************************(((.((.((((((....(((.((((...................(((.((((.(((((((((..........(((((....)))))......))))))))).)))).))).......)))).))).....)))))).........)).)))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M044 female body |
V055 head |
M058 embryo |
V105 male body |
V106 head |
|---|---|---|---|---|---|---|---|---|---|---|
| ......................................CGTAATGATTATTAGGGAC.................................................................................................................................................................................................. | 19 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................................CGTAATGATTAATTGGGAC.................................................................................................................................................................................................. | 19 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................................CCGTAATGATTAATAGGGAC.................................................................................................................................................................................................. | 20 | 3 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................TTTATGGTCGAAACTTCG........... | 18 | 2 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...............ATGAATTAAGTGTTGTTCTTG....................................................................................................................................................................................................................... | 21 | 3 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................ATCGTAATGATTATTAGGGAC.................................................................................................................................................................................................. | 21 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..............................................................................................................TAACCTTGCAGTTAGGTAGCAGCTTA................................................................................................................... | 26 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...........................................TGATTATGTGGGACGTGTAT............................................................................................................................................................................................ | 20 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................TACCCTTGCAGTTAGGTAGCAG....................................................................................................................... | 22 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................................................................................................................................TCTCCGGTGCCGATCATCG..................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| .......................................GTAATGATTATTAGGGAC.................................................................................................................................................................................................. | 18 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
|
TGTATCGGGCGTTAGTACTTAATTCACTATAAAAACTGGTATTACTAATAAACCCTGAAAATAAAAAAGCGAGTCTATTATTATTACTAATAAAGCCTGGCTTCACATATATGGGAACGTCAATCCATCGTCGAATATTAAAATATATATATAGCCTAGCATATATCGACTGGCTAGGAATACTCTTAAAGTAGTAGTGTCCACGGCCAGTAGCGTGTCCAAAAATACGTGCTTTGAAGCGTCAAGCGCTG
**************************************************(((.((.((((((....(((.((((...................(((.((((.(((((((((..........(((((....)))))......))))))))).)))).))).......)))).))).....)))))).........)).)))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M058 embryo |
V105 male body |
M044 female body |
V039 embryo |
|---|---|---|---|---|---|---|---|---|---|
| ......................................................................................................................................................................................................GTCCACGGCCAGTAGCGTGTCCAAA............................ | 25 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ........................................................GAGAAAAAACAAGCGAGTCTA.............................................................................................................................................................................. | 21 | 3 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 |
| .................................................................................................................GGAACGTCAATCCATCGTCGAATAT................................................................................................................. | 25 | 0 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 |
| TGTATCGGCCGTTAGAAC......................................................................................................................................................................................................................................... | 18 | 2 | 16 | 0.13 | 2 | 2 | 0 | 0 | 0 |
| TGTATCGGCCGTTAGAACA........................................................................................................................................................................................................................................ | 19 | 3 | 20 | 0.10 | 2 | 1 | 0 | 1 | 0 |
| ..............................................................................................................................................................GCATATATCGACTGGCTAGGAA....................................................................... | 22 | 0 | 10 | 0.10 | 1 | 0 | 0 | 0 | 1 |
| ............................................................................................................ATATGGGAACGTCAATCCATCGTCGA..................................................................................................................... | 26 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................ACCGTCAGTAGCGTGTCCC.............................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| .............................................................................................................TATGGGAACGTCAATCCATCGTCGAAT................................................................................................................... | 27 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| .....................................................................................................................CGTCAATCCATCGTCGAATA.................................................................................................................. | 20 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................TACTCTTGGAGTAGTAGT..................................................... | 18 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ................................................................................................................GGGAACGTCAATCCATCGTCGAAT................................................................................................................... | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................CATAGCCTAGCATATATCGAC................................................................................. | 21 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ...............................................................................................................TGGGAACGTCAATCCATCGTCGAACAT................................................................................................................. | 27 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ..............................................................................................................ATGGGAACGTCAATCCATCGTCGAATAT................................................................................................................. | 28 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droAna3 | scaffold_13499:218008-218258 - | dan_3362 | ACATAGCCCGCAATCATGAATTAAGTGATATTTTTGACCATAATGATTATTTGGGACTTTTATTTTTTCGCTCAGATAATAATAAT-GA------------------TTATTTCGG-----ACCGAAGTGTATATACCCTTGCAGTTAGGTAGCAGCTTATAATTTTATATATATATCGG--ATCGTATATAGCTGACCGATCCTTATGAGAATTTCATCATCACAGGTGCCGGTCATCGCACAGGTTTTTATGCACGAAACTTCGCAGTTCGCGAC |
| droBip1 | scf7180000395478:41240-41416 + | CCATAGCGCGAAATCATGAATTGAGTGATATTTTTGAGCAAAA--------------------------------ATAATAAAAAT-CAAAAACAAGAAAGGAAAGCTAACTTCGGGCGGAGCCGAAGTTGATATACCCTTGCAGTTA------------AAACCGGATATATATCACAAATATCGGATATAGTTGGCCGATCCTTATAAGAATATGATAAT------------------------------------------------------- | |
| droFic1 | scf7180000454069:158277-158359 + | GGTTATCTAAAAA--------------------------------------------CTTTATTTTAACT----GATA--------------ACAAGAAAGGA-AGCCAGCTTCGGCA--AGCCGAAGCTTATATACCCTTGCAGTCA--------------------------------------------------------------------------------------------------------------------------------- | |
| droEle1 | scf7180000489338:948-1043 + | AAATAAATCGAAAAAATAAATAAAA--AAAATTATAACTTTGCTGTTTATTAATTTTTTTTAGTTCTTCGA-CATATAGTAATGGTTAA------------------ATATTTCAG-----A----------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droRho1 | scf7180000778355:6019-6105 + | CCT------GCAATC--GACTTTAGTGAGTTTTTTGA-------------------------GTTTTTCCC-T------------------------CCAGGA-AGCAAACTTCGGCC--AGCTGAAGTTTATATACCCTTGCAGTTA--------------------------------------------------------------------------------------------------------------------------------- | |
| droBia1 | scf7180000301495:203527-203563 + | ACT-------------------------------------------------------------------------------------------------------------TCGGCA--AGCCGAAGTTTGTATACCCTTGCAGTTA--------------------------------------------------------------------------------------------------------------------------------- | |
| droSec2 | scaffold_87:94286-94311 + | GA-----------------------------------------------------------------------------------------------------------------------ACCGAAGTTTATATACCCTTGCAG------------------------------------------------------------------------------------------------------------------------------------ | |
| dp5 | Unknown_singleton_3462:99-125 - | GGT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCTGGTCTTCGCACAGGTTTTTAT------------------------ | |
| droPer2 | scaffold_121:43703-43733 - | GGT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCTGGTCATCGCACAGGGTTTTG-GCACG------------------- | |
| droMoj3 | scaffold_1874:5671-5673 + | GAT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
Generated: 05/18/2015 at 07:14 AM