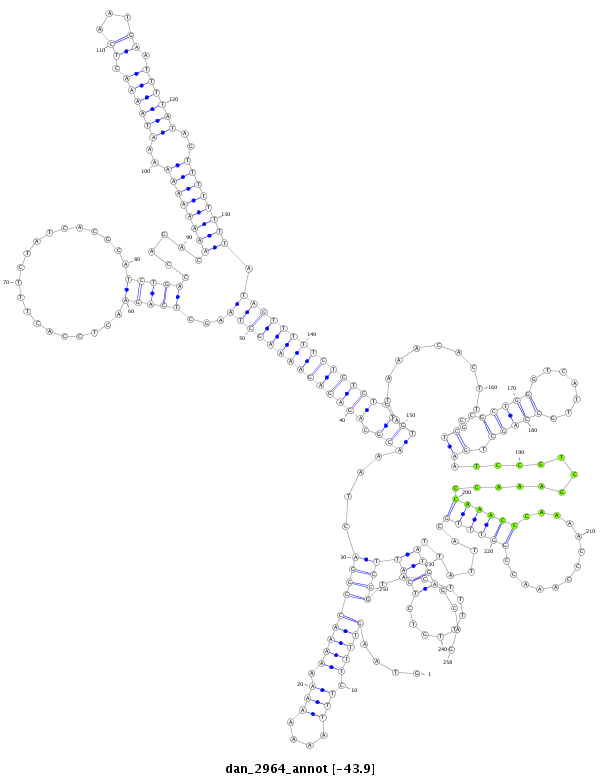
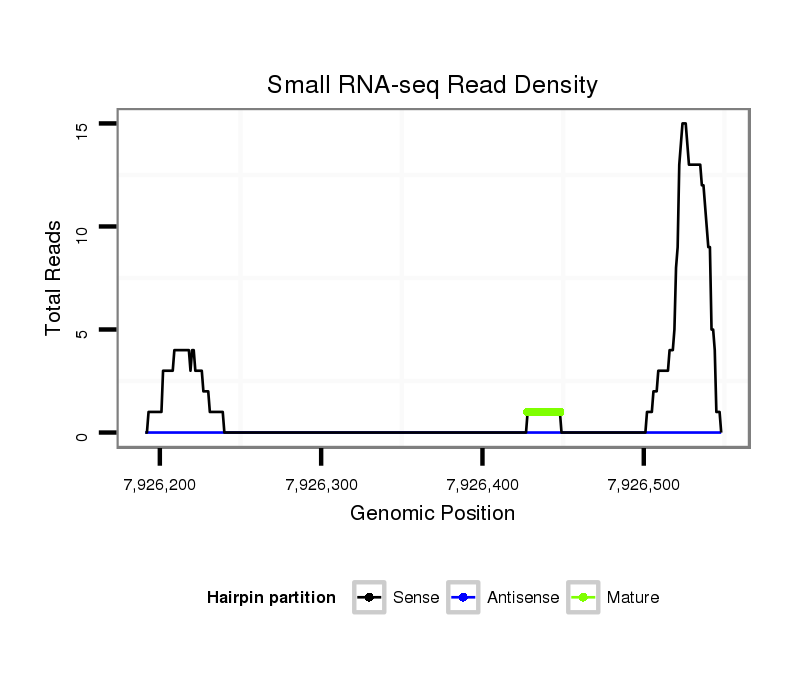
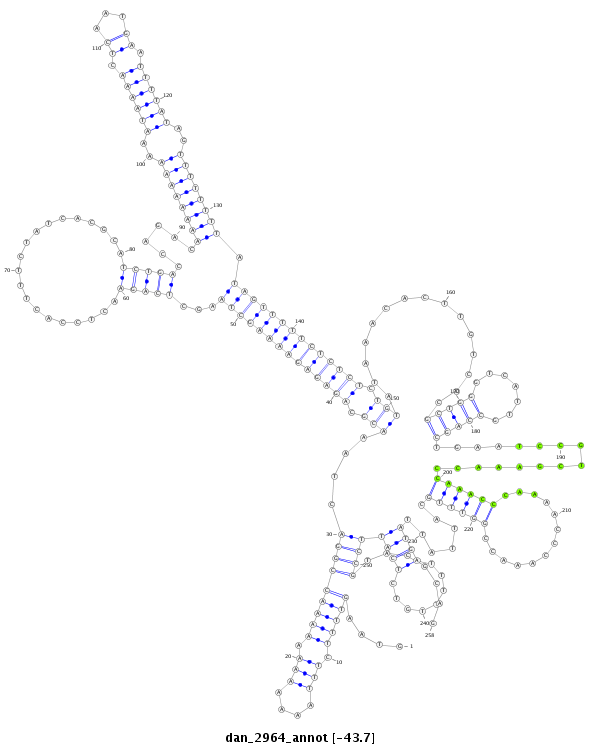
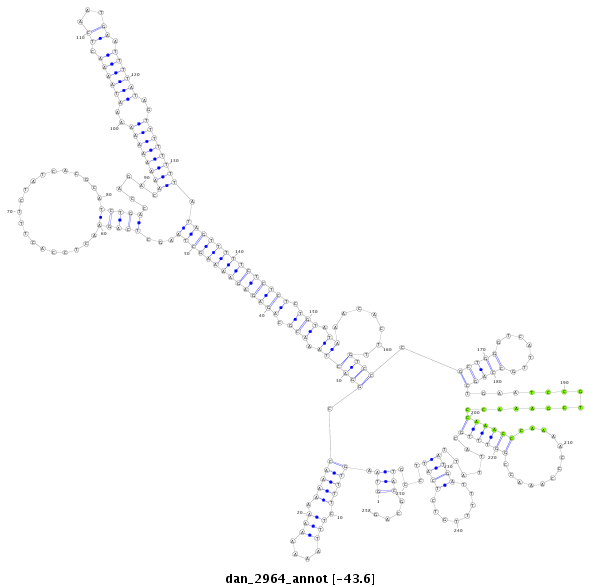
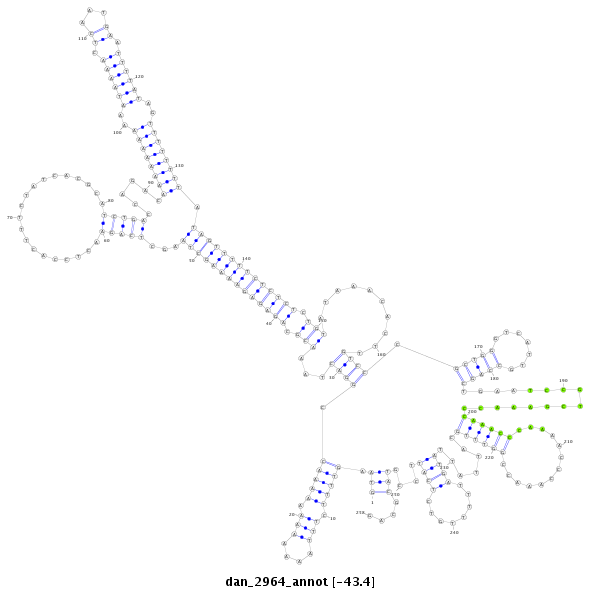
ID:dan_2964 |
Coordinate:scaffold_13340:7926241-7926498 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -43.7 | -43.6 | -43.4 |
 |
 |
 |
CDS [Dana\GF17989-cds]; CDS [Dana\GF17989-cds]; exon [dana_GLEANR_19249:1]; exon [dana_GLEANR_19249:2]; intron [Dana\GF17989-in]
No Repeatable elements found
| ##################################################------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## GAGCCCGGAAACTACAAACGCGCCACAAAGCGAATCGAAGATGGCTACAAGTAAGTTTTCTTTAAAAAAAAAAAACCGGACTAAACGCAGAGAGAAAAGCTAAGCTCAGAACTCCACTTTCTATCACGCATCTGACCAGACAAAAAAAAAAATAAAACTCAATGAATTTTATAGTTTTTTTTATAGTTTTTCTCTCTCTGTATAAACACTTGTCCCGCTGGGTCATTGCCAGCTGAATCCGTCGAAACCCAAACCCAAAACCCAAACCGGTTTGCATTATTATGATTTTTGTCTCATTCCGTACGCAGACTCTGCAATGACCTACAGCAACTGGTGCAGGAGCGAGCCGACATCGAGA **************************************************....(((((.(((....))).)))))((((....(((.((((((((((((((...(((((....................)))))......((((((((..((((((.((...)).))))))..)))))))).)))))))))))))).)))...........((..(((((.......))))).)).............((((((.............)))))).......((((........)))))))).......************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V106 head |
M058 embryo |
M044 female body |
V055 head |
V105 male body |
V039 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................................................................................................................................................................................................TGGTGCAGGAGCGAGCCGAC....... | 20 | 0 | 1 | 3.00 | 3 | 0 | 1 | 1 | 0 | 1 | 0 |
| ...........CTACAAACGCGCCACAAAGC....................................................................................................................................................................................................................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................GGTGCAGGAGCGAGCCGACATC.... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................ACTGGTGCAGGAGCGAGCGG......... | 20 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................ACTGGTGCAGGAGCGAGCCGACATC.... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................GGTGCAGGAGCTAGCCGACATCGAGA | 26 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................AACTGGTGCAGGAGCGAGCCGACAT..... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................................................................................................................GACCTACAGCAACTGGTGCAGGAGCGA............. | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................................................................................................................................................................................TCCGTCGAAACCCAAACCCAA.................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................AATGACCTACAGCAACTGGTGC..................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..GCCCGGAAACTACAAACGCGCCACAA.......................................................................................................................................................................................................................................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .......................................................................................................................................................................................................................................................................................................................CTGCAATGACCTACAGCAACTGGTG...................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................ACTGGTGCAGGAGCGAGCCG......... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................................................GTGCAGGAGCGAGCCGACATCGAG. | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........CTACAAACGCGCCACAAAGCGAATC.................................................................................................................................................................................................................................................................................................................................. | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................CGCGCCACAAAGCGAATCGAAG.............................................................................................................................................................................................................................................................................................................................. | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................GACCAGACAAAAGAAAAAA.............................................................................................................................................................................................................. | 19 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................GCGAATCGAAGATGGCTACA..................................................................................................................................................................................................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................................................................................................................................GGTGCAGGAGCGAGCCGG........ | 18 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................................AGCAACTGGTGCAGGAGCGAGC........... | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................................TGGTGCAGGAGCGAGCCGACATC.... | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................ACTGGTGCAGGAGCGAGCC.......... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................CTGGTGCAGGAGCGAGCCGAC....... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ....................................................................................................................................................................................................................................................AAACCCCTACCCAAAAACCAAACC.......................................................................................... | 24 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..GCCCGGAAACTACCAACTCC................................................................................................................................................................................................................................................................................................................................................ | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................................GCAGGAGCAAGCCGACTTC.... | 19 | 2 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................AAAAAAAAAAACCCGGAC..................................................................................................................................................................................................................................................................................... | 18 | 1 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................GAAAACCAAACCCAACACCCCA............................................................................................. | 22 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...................................................................................................................................CTGACAAGACAGAAAAAAAAAGA............................................................................................................................................................................................................ | 23 | 3 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......GAAACTACAAAGGAGCCACCA.......................................................................................................................................................................................................................................................................................................................................... | 21 | 3 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................................AAAAAAAAACACCGGACCAA.................................................................................................................................................................................................................................................................................. | 20 | 2 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................ACGCGCATCTGATCAGACA........................................................................................................................................................................................................................ | 19 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...............................................................................................................................................................................................................................................CGACGACACCCAAACCAAAAA.................................................................................................. | 21 | 3 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................AAAAAATAAAACTCAAAGA................................................................................................................................................................................................. | 19 | 1 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...................................................................................................................................................................................................................................................GAAACCCAAGCCCCAAACCC............................................................................................... | 20 | 2 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................AAAAAAAACACCGGACCAA.................................................................................................................................................................................................................................................................................. | 19 | 2 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................GAAACCCAGACCCCAAACC................................................................................................ | 19 | 2 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................AAAAAAAAAAATTAAACTCAATAA................................................................................................................................................................................................. | 24 | 2 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..............................................................AAAAAAAAAAAAACCGGAC..................................................................................................................................................................................................................................................................................... | 19 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................AAAAAAAAAAATAAACCTC...................................................................................................................................................................................................... | 19 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................ACTGGCGGAGGAGCGAGC........... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................................................................................................................................................................................................................................................CGAACACCAAACCCAAAAC................................................................................................. | 19 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................AAAAAAAACAGGACGAAACCCA............................................................................................................................................................................................................................................................................. | 22 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................TTATAGTTTTTTTGATGG............................................................................................................................................................................ | 18 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
|
CTCGGGCCTTTGATGTTTGCGCGGTGTTTCGCTTAGCTTCTACCGATGTTCATTCAAAAGAAATTTTTTTTTTTTGGCCTGATTTGCGTCTCTCTTTTCGATTCGAGTCTTGAGGTGAAAGATAGTGCGTAGACTGGTCTGTTTTTTTTTTTATTTTGAGTTACTTAAAATATCAAAAAAAATATCAAAAAGAGAGAGACATATTTGTGAACAGGGCGACCCAGTAACGGTCGACTTAGGCAGCTTTGGGTTTGGGTTTTGGGTTTGGCCAAACGTAATAATACTAAAAACAGAGTAAGGCATGCGTCTGAGACGTTACTGGATGTCGTTGACCACGTCCTCGCTCGGCTGTAGCTCT
**************************************************....(((((.(((....))).)))))((((....(((.((((((((((((((...(((((....................)))))......((((((((..((((((.((...)).))))))..)))))))).)))))))))))))).)))...........((..(((((.......))))).)).............((((((.............)))))).......((((........)))))))).......************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V055 head |
M044 female body |
V106 head |
V039 embryo |
V105 male body |
|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................................................................................................................................................................................................................................................................................TCCTTTCTCGGCTGTAGCTGT | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................................................................CTACTCTCTCGGCTGTAGCTC. | 21 | 3 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................................CCTGCTCGGCTGTAGCTC. | 18 | 2 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................TCGATTAGTTTCTACCGA........................................................................................................................................................................................................................................................................................................................ | 18 | 2 | 9 | 0.11 | 1 | 0 | 0 | 1 | 0 | 0 |
| .........................................................................................................................................................................................................................................................GTTTGGGTGCTGGGGTTGG.......................................................................................... | 19 | 3 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 |
| ...................................................................................................GAGTCGAGTCTTGAGCTG................................................................................................................................................................................................................................................. | 18 | 2 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................AGCTTTGGGTTTGGGTTGA.................................................................................................. | 19 | 2 | 14 | 0.07 | 1 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................................................................TCCGTAGACTCGTCTGTTG...................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| ..................................................................................................................................................................................................................................................................................................................................................CCTTCCTCGGCTGTAGCTT. | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droAna3 | scaffold_13340:7926191-7926548 + | dan_2964 | GAGCCCGGAAACTACAAACGCGCCACAAAGCGAATCGAAGATGGCTACAAGTAAGTTTTCTTTAAAAAAAAAAAACCGGACT---AAACGCA--------------GAGAGAAAAGCTAAGCTCAGAACTCC-ACTTTCTATCACGCATC-------TGACCAGACAAAAAAAAAAATAAAACTCAATGAATTTTATAGTTTTTTTTATAGTTTTTCTCTCTCTGTAT------AAACACTTGTC-CCGCTGGGTCATTGCCAGC-------------TGAATCCGTCGAAACCCAAA-CCCA-------------------------------AAA--C-----------CCAAACCGGTT------------------------------TGCATTATTATGATTTT-------TGTC---------------------------------------TCATTCCG---------------------------------------T---ACGCAGACTCTGCAATGACCTACAGCAACTGGTGCAGGAGCGAGCCGACATCGAGA |
| droBip1 | scf7180000396708:920973-921327 - | GAGCCCGGAAACTATAAACGCGCCACAAAGCGAATCGAAGATGGATACAAGTGAGTTCCCATTAAAA-------------CTATAAAACCCA--------------GAGAGAAAATCTAAAATCAGAACTAAAACTTTCTATCATGAATCTCATAAATGACCA--------------------------------------TTTTGTTTAGTTTTTCTCTCTGTGTACTCTCTCTAACACTTGTC-CCGCTGGGTCATTGCCAGC-------------TGAATCCGTCGAAACCCATA-CCCATACTCATACCCATACCCATATCCGAACCAAAAAA--C-----------CCAAACCGGTT------------------------------TACATTATTTTGATTTT-------TGTC---------------------------------------TCATTCCG---------------------------------------T---ACGCAGACTCTGCAATGACCTACAGCAACTGGTGCAGGAGCGGGCCGACATCGAGA | |
| droKik1 | scf7180000302307:90319-90496 - | T---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTTCTCTGTGCAT------AAGCACTTGGT-CCCCTGGGTCATTGCCAGCGACAGCAGCAGGCAGAGTCACTCGGAAAGC---------------------------------------AAA--G-----------CCAAACCGGTT------------------------------CGCATTATTTTGATTTC-------TGTC-TC------------------------------------TCGTCTCG---------------------------------------C---ACGCAGACTCTGCAATGACCTACAGCAACTGATACAGGAGCGAGCCGACATCGAGA | |
| droFic1 | scf7180000453930:117087-117267 - | T---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTGCTCTGTGTAT------AAGCACTTGGC-TCGCTGGGTCATTGCCAGC-------------AGAATCCATTCGAACCCGAA-CCCAACCCCAAACCC--------------------AAA--T-----------CCAAACCGGTT------------------------------TGCATTATTTTGATTTT-------TGTC---------------------------------------TCATTTCA---------------------------------------C---ACGCAGACTCTGCAATGACCTACAGCAACTGATTCAGGAGCGTGCCGAAATCGAGA | |
| droEle1 | scf7180000491080:540896-541076 + | T---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTTCTCTGTGTGT------AAGCACTTGGC-CCTCTGGGTCATTGCCAGC-------------AGAATCCATCGAAACCCGAA-TCCAAACCCAAATAC--------------------AAG--C-----------CCAAACCGGTT------------------------------TGCATTATTATGATTTT-------TGTC---------------------------------------TCATTTCG---------------------------------------C---ACGCAGACTCTGCAATGACCTACAGCAACTGATTCAGGAGCGTGCCGACATCGAGA | |
| droRho1 | scf7180000779659:242446-242620 + | TT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTTCTCTGTGCAT------AAGCACTTGGC-CCGCTGGGTCATTGCCAGC-------------AGAATCCATCTGAACCCGAA-CCCAAATAC--------------------------AAA--C-----------TCAAACCGGTT------------------------------TGCATTATTATGATT-T-------TGTC---------------------------------------TCATTTCG---------------------------------------C---ACGCAGACTCTGCAATGACCTACAGCAACTGATTCAGGAGCGTGCCGACATCGAGA | |
| droBia1 | scf7180000302402:9500922-9501097 + | T---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTTCTCTGTGCAC------CAAGACTTGGA-CCTCTGGGTCATTGCCAGC-------------TGAATCCATCGGAACGCGAA-CCCAGGCCCAA-------------------------AT--C-----------CAAAACCGGTT------------------------------TGCATTATTGTGATTTT-------TGTC---------------------------------------TCATTTCG---------------------------------------C---CCGCAGACTCTGCAATGACCTACAGCAACTGATACAGGAGCGTGCCGAAATCGAGA | |
| droTak1 | scf7180000415245:48348-48529 - | TT------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GATAGT--TTTTCTCTGTGCAC------GAGCACCTGGA-CCGCTGGGTCATTGCCAGC-------------TGAATCCATCGGAACTCGAA-CCCAAACCCA--------------------------AA--A-----------CCAAACCGGTT------------------------------TGCATTATTATGATTTT-------TGTC---------------------------------------TCATTTCA---------------------------------------C---CCGCAGACTCTGCAATGACCTACAGCAACTGATACAGGAGCGTGCCGAAATCGAGA | |
| droEug1 | scf7180000409542:519705-519884 - | A---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TAGTT-TTTTTACTGTGTAT------TACCACTTGGC-CTGCTGGGTCATTGCCAGC-------------TCAATCCATCGCAACCCGAA-CCCAAACCA--------------------------ATA--A-----------TCAAACCGGTT------------------------------TGCATTATTATGATTTT-------TGTC---------------------------------------TCATTTCG---------------------------------------C---CCGCAGACTCTGCAATGACCTACAGCAACTGATACAGGAGCGTGCCGACATGGAGA | |
| dm3 | chr3R:16661464-16661646 + | T----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCTCTCTGTGTAT------TTGCACTGGGCGACGCTGGGTCATTGCCAGC-------------CGAATATATCGACACCCGAAGCCCAAACCCAAGACA-------------------AAAA--T-----------TCAAACCGGTT------------------------------TGCATTATTATGATTCT-------TGTC---------------------------------------TCATTTCG---------------------------------------C---CCGCAGACTCTGCAATGACCTACAGCAACTGATTCAGGAGCGTGCCGACATCGAGA | |
| droSim2 | 3r:4643952-4644133 - | T----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCTCTCTGTGTAT------TTGCACTGGGCGACGCTGGGTCATTGCCAGC-------------CGAATATATCGGCACCCGAA-CCCAAACCCAAGCCA-------------------AAAA--T-----------CCAAACCGGTT------------------------------TGCATTATTATGATTCT-------TGTC---------------------------------------TCATTTCG---------------------------------------C---CCGCAGACTCTGCAATGACCTACAGCAACTGATTCAGGAGCGTGCCGACATCGAGA | |
| droSec2 | scaffold_27:73574-73767 - | TC---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTTTAAGG--TTCTCTCTGTGTAT------TTGCACTGGGCGACGCTGGGTCATTGCCAGC-------------CGAATATATCGGCACCCGAACCCCAAACCCAAGCCC-------------------AAAT--T-----------TCAAACCGGTT------------------------------TGCATTATTATGATTCT-------TGTC---------------------------------------TCATTTCG---------------------------------------C---CCGCAGACTCTGCAATGACCTACAGCAACTGATTCAGGAGCGTGCCGACATTGAGA | |
| droYak3 | 3R:5458283-5458466 + | AG-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTCGT--TTCTCCATGTGTAT------T-GCACTGGGCGACGCTGGGTCATTGCCAGC-------------CGAGTATATCGGCACCCGAA-CCAAAACCCAAGCCC-------------------AAAA--G-----------CCAAACCGGTT------------------------------TGCATTATTATGATTCT-------TGTC---------------------------------------TCATTC----------------------------------------------CGCAGACTCTGCAATGACCTACAGCAACTGATACAGGAGCGTGCCGACATAGAGA | |
| droEre2 | scaffold_4770:4994786-4994967 - | T---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTGTCTCTGTGTAT------T-GCACTGGGCGACGCTGGGTCATTGCCAGC-------------CGAGTATAAAGTCAACCGAA-CCAAAACCCAAACCC-------------------AAAA--G-----------CCAAACCGGTT------------------------------CGCATTGTTATGATTCT-------TGTC---------------------------------------TCATTTCG---------------------------------------C---CCGCAGACTCTGCAATGACCTACAGCAACTGATACAGGAGCGTGCCGACATCGAGA | |
| dp5 | 2:12782350-12782442 - | TG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTTTATTAT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------TT------ATTTC-------TGTTCTCTTT---------------------------------CCCTCTCT---------------------------------------T---GCGCAGACTCTGCAATGACCTACAGCAACTGATTCAGGAGCGAGCCGACATCGAAA | |
| droPer2 | scaffold_0:7676346-7676438 + | TG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTTTATTAT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------TT------ATTTC-------TGTTCTCTTT---------------------------------CCCTCTCT---------------------------------------T---GCGCAGACTCTGCAATGACCTACAGCAACTGATTCAGGAGCGAGCCGACATCGAAA | |
| droWil2 | scf2_1100000004921:2201739-2201814 - | TT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CTTT---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTATTTT-------TGTT---------------------------------------T------T---------------------------------------C---TGACAGACTCTGCAATGACCTACAGCAACTGATACAGGAGCGAGCTGATATTGAGA | |
| droVir3 | scaffold_13047:12048669-12048725 - | T-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCACAGACTCTGCAACGACCTACAGCAACTGATACAGGAGCGCGCCGATATTGAGA | |
| droMoj3 | scaffold_6540:29731437-29731677 + | GAGCCCGGCAATTACAAACGAACCACCAAACGCATCGAAGACAGCTACAAGTGAGT----------------------------ATG----AAAATACAAATCGTAGA--------------------------------------------------------------------------------------------------------------CACTG-------------------------TTAGGGTCATTGCCAGC-------------AATAACAAACTGG-------------------------------------------------------------AAAACCAGTTATT---------------------------TGT---------ATTGT-------CGCTCTCGCTCACTCTCTCTGTGTGTCTTTCCTTCTCGCTTTCTCATCT-G---------------------------------CC----C---GCACAGACTCTGCAATGACCTACAGCAACTGATACAGGAGCGTGCCGATATTGAGA | |
| droGri2 | scaffold_14830:1163437-1163763 + | GAGCCTGGCAATTATAAACGAACCACCAAACGCATCGAAGATAGCTACAAGTGAGT----------------------------AAAACACAAAATACAAATCGCAAATCAAAACACAA---------------------------------------------------------------------------------------------------CACT---------------GGCTTGGC-TGCTTGGGTCATTGCCAGC-------------AATAACAA------------------AAACAA-----------------------AAAAAAGTGTACAACTTAAATAACCGGTTATTTACTTGTCTCTTTTCTTTTCTTTATTTTGT---------ATTCTCGCTCTCTCTTCTCTGC---------------------------------CCCTCTCTCTATTTATATTTGATGGCTCATCCACACATCTGTCGGTGCTCGGCACAGACTCTGCAATGACCTACAGCAACTGATACAGGAGCGCGCCGATATTGAGA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/18/2015 at 05:57 AM