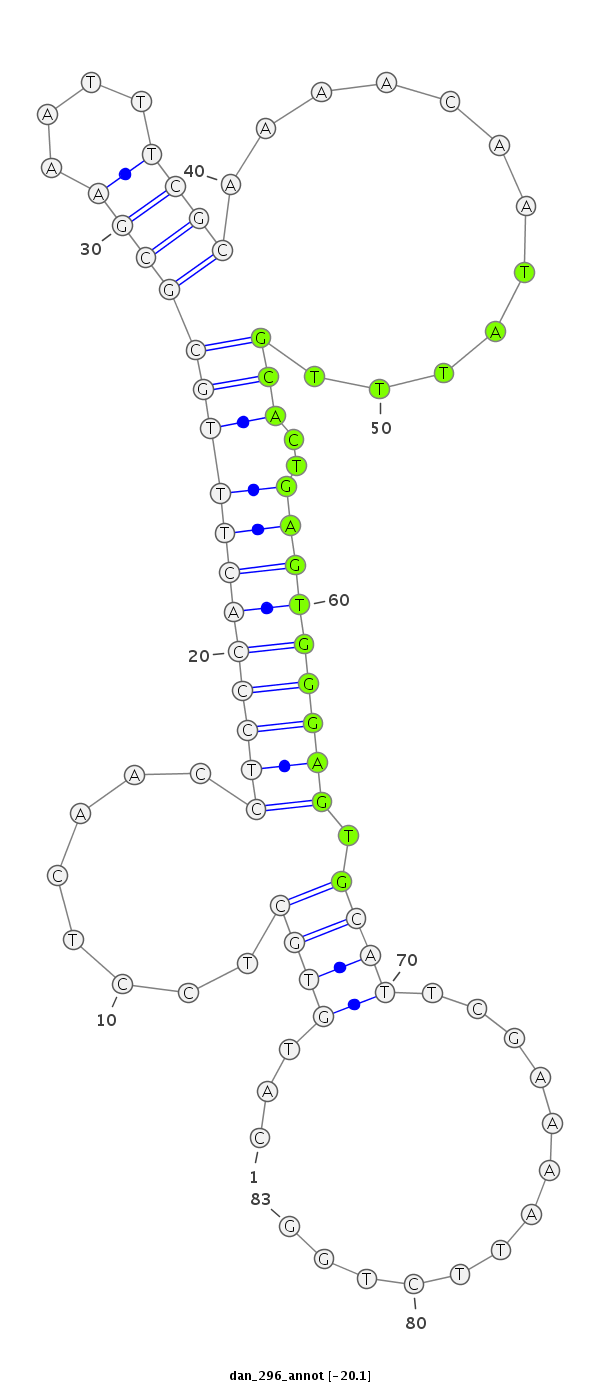
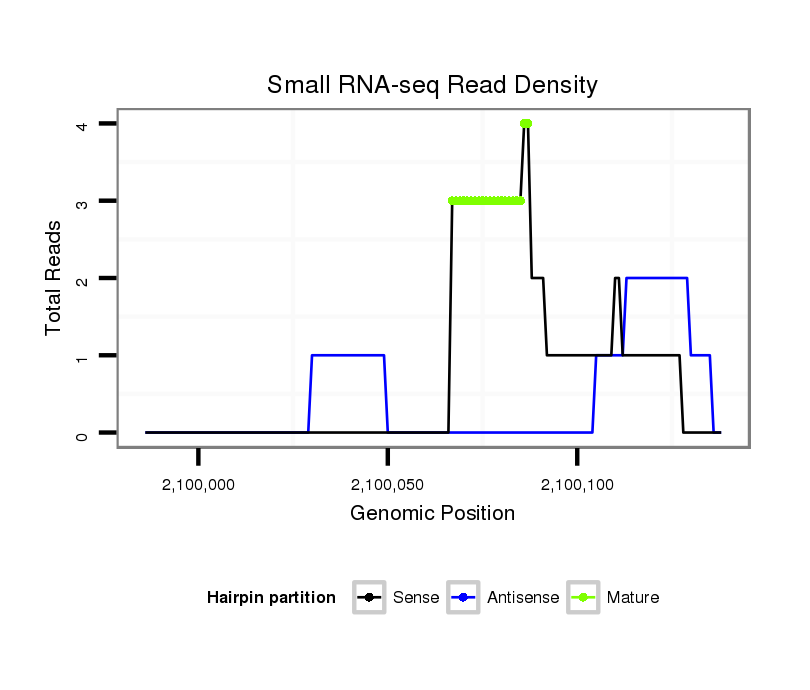
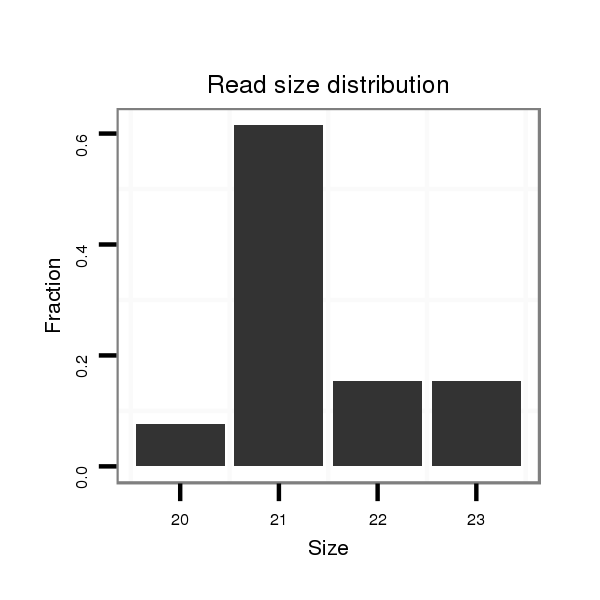
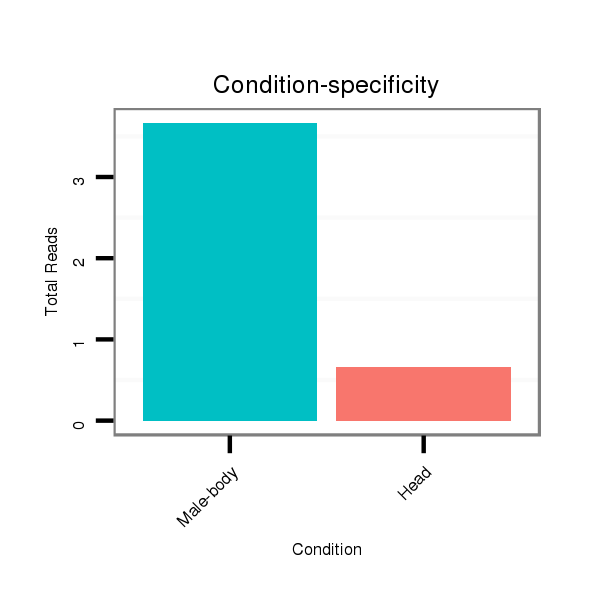
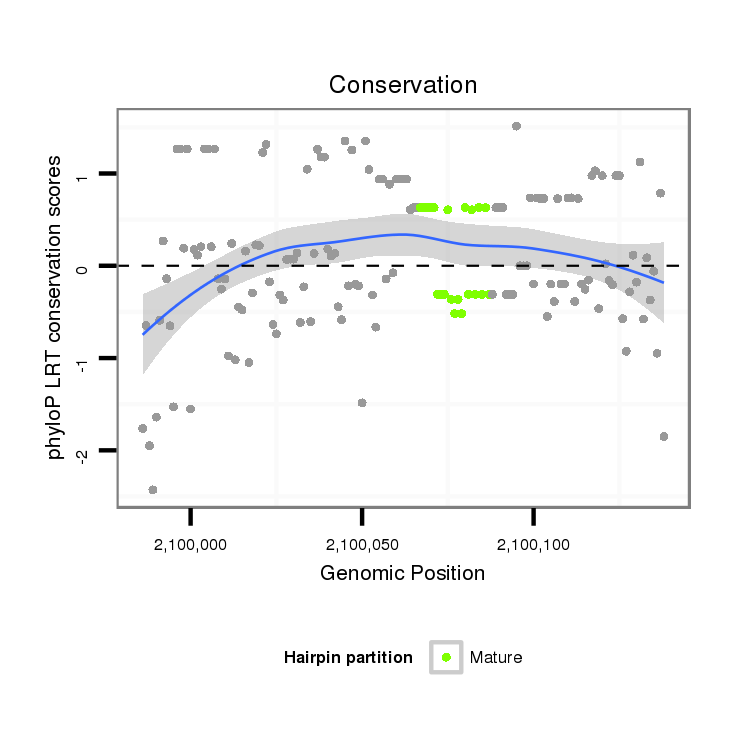
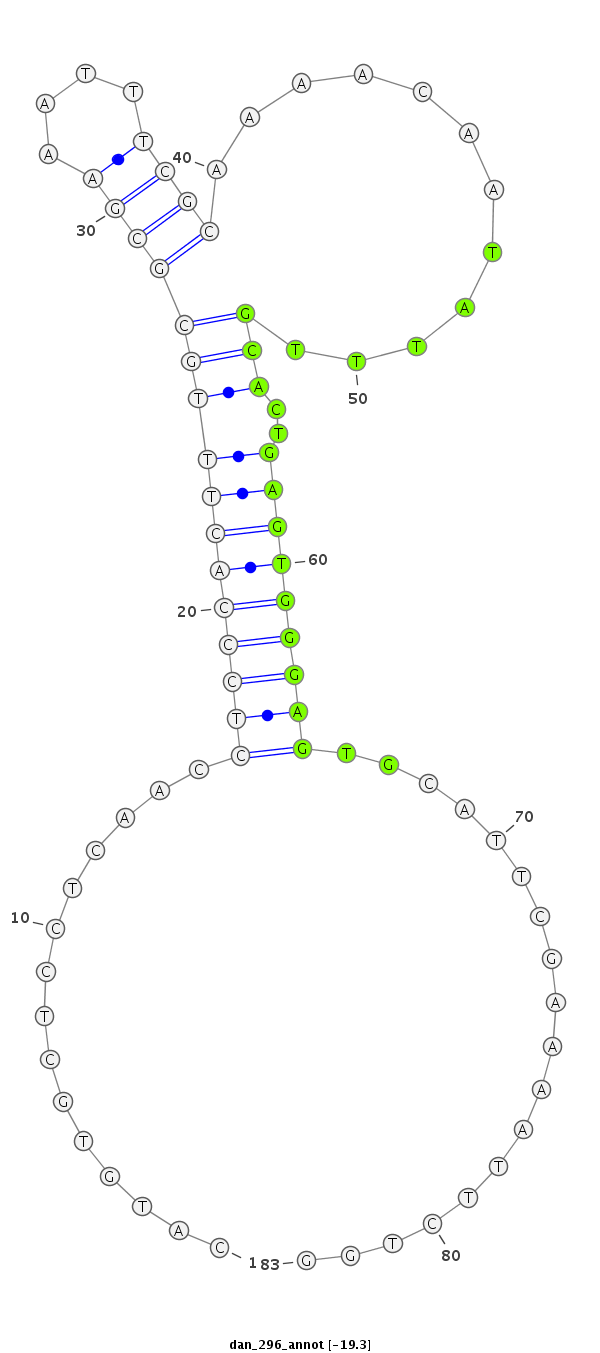
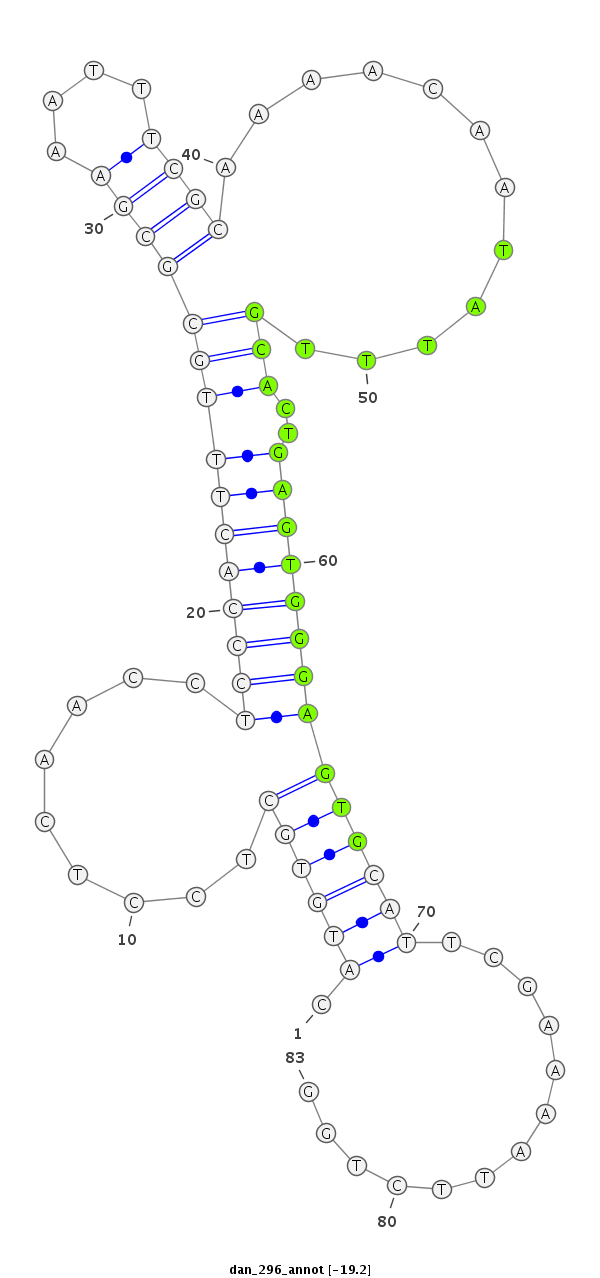
ID:dan_296 |
Coordinate:scaffold_12911:2100036-2100088 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -19.3 | -19.2 |
 |
 |
intergenic
No Repeatable elements found
| mature | star |
|
CACGGAAACGTTTTCGGAATTTGGAGACATGTCTCCATGTGCTCCTCAACCTCCCACTTTGCGCGAAATTTCGCAAAACAATATTTGCACTGAGTGGGAGTGCATTCGAAAATTCTGGGCTCAATTCCGTCGTCCCTCGGACCTTCTGGAATC
***********************************...((((........((((((((((((((((....))))............)))..))))))))).)))).............*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V105 male body |
M058 embryo |
M044 female body |
V106 head |
V039 embryo |
V055 head |
|---|---|---|---|---|---|---|---|---|---|---|---|
| .................................................................................TATTTGCACTGAGTGGGAGTG................................................... | 21 | 0 | 3 | 2.33 | 7 | 5 | 0 | 0 | 2 | 0 | 0 |
| .................................................................................TATTTGCACTGAGTGGGAGTGCATT............................................... | 25 | 0 | 3 | 1.00 | 3 | 1 | 1 | 0 | 1 | 0 | 0 |
| ....................................................................................................TGCATTCGAAAATTCTGGGCTCAATT........................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................................TTCCGTCGTCCCTCGGAC........... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................ATGTCTACATGAGCTCCAC.......................................................................................................... | 19 | 3 | 13 | 0.69 | 9 | 0 | 8 | 0 | 0 | 1 | 0 |
| ...............................................................................AATATTTGCACTGAGTGGGAGTG................................................... | 23 | 0 | 3 | 0.67 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................TATTTGCACTGAGTGGGAGTGC.................................................. | 22 | 0 | 3 | 0.67 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................ATCAGGGCTCAATTCCGT....................... | 18 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..............................................................................................................................CCGTCGTCCCTCGGACCTTCTGGAATC | 27 | 0 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................................AATCTGGGCTCGATTCCG........................ | 18 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ............................................................................................................................TTCCTTCGTCCCTCGGACCTTCTGG.... | 25 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................ATGTCTACATGAGCTCCCCA......................................................................................................... | 20 | 3 | 7 | 0.43 | 3 | 0 | 2 | 0 | 0 | 1 | 0 |
| ............................ATGTCTACATGAGCTCCACA......................................................................................................... | 20 | 3 | 7 | 0.43 | 3 | 0 | 1 | 0 | 0 | 2 | 0 |
| ............................ATGTCTACATGAGCTCCCC.......................................................................................................... | 19 | 3 | 12 | 0.42 | 5 | 0 | 4 | 0 | 0 | 1 | 0 |
| .................................................................................TATTTGCACTGAGTTGGAGTG................................................... | 21 | 1 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................TTTGCACTGAGTGGGAGTGCAT................................................ | 22 | 0 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................TATTTGCACTGAGTGTGAGTG................................................... | 21 | 1 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................ATCTATTTGCACTGAGTGGGAGTGCATT............................................... | 28 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................TATTTGAACTGAGCGGGAGTGCATT............................................... | 25 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................TATTTGCACTGAGTGGGAGT.................................................... | 20 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................TATTTGCACTGAGTCGGAGTG................................................... | 21 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................CAACATTTGCACTGAGTGGGA...................................................... | 21 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................CAATATTTGCACTGAGTGGGA...................................................... | 21 | 0 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................CGATTTGCACTGAGTGGGAGTGC.................................................. | 23 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................AGTGGGAGGGCATTCGAC........................................... | 18 | 2 | 11 | 0.09 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..................................................................................................................CGGGGTTCAATTCCGTCGG.................... | 19 | 3 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................ATTCGCGCTGAGCGGGAGT.................................................... | 19 | 3 | 12 | 0.08 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................TCAATTCCGTAGTGACTCG.............. | 19 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| .....AAACGTGGTCGGAATCTGG................................................................................................................................. | 19 | 3 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................CAGGGCTCAATTCCGGAG..................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...........................................................................CAACAATATTTGCACTGC............................................................ | 18 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
|
GTGCCTTTGCAAAAGCCTTAAACCTCTGTACAGAGGTACACGAGGAGTTGGAGGGTGAAACGCGCTTTAAAGCGTTTTGTTATAAACGTGACTCACCCTCACGTAAGCTTTTAAGACCCGAGTTAAGGCAGCAGGGAGCCTGGAAGACCTTAG
***********************************...((((........((((((((((((((((....))))............)))..))))))))).)))).............*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M044 female body |
M058 embryo |
V055 head |
V039 embryo |
V106 head |
|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................................................................GAGTTAAGGCAGCAGGGAGCCTGGA......... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................GAGTTGGAGGGTGAAACGCG......................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................GCAGCAGGGAGCCTGGAAGACCT... | 23 | 0 | 2 | 1.00 | 2 | 1 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................GCAGCAGGGAGCCTGGAAGACCTT.. | 24 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................TGTATACGAGGACTTGGAGGGT................................................................................................. | 22 | 3 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................................................GGCAGCAGGGAGCCTGGAAGACCTT.. | 25 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................GTTTTGTTGTAAACGTGATTCC.......................................................... | 22 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..............................................................................................................................GGCAGCAGGGAGCCTGGAAGACCT... | 24 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................................TTGCAGGGAGCCTGGAAGACCTT.. | 23 | 2 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 |
| GTGACTTTACAAAAGCCATAA.................................................................................................................................... | 21 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 |
| ........................................CGAGGATTAGGAGGGTCAA.............................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ........................................CGAGGACTTGGAGGGATA............................................................................................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| .............................................................................TGATATAAACGTGTCTCGC......................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droAna3 | scaffold_12911:2099986-2100138 + | dan_296 | CACGGAAACGT------TTTCGGAATTTGGAGACATGTCTCCATGTGCTCCTCAACCTCCCACTTTGCGCGAAATTTCGCAAAACAATATTTGCACTGAGTGGGAGTGCATTCGAAAATTCTGGGCTCAATTCCGTCGTCCCTCGGACCTTCTGGAATC |
| droKik1 | scf7180000302696:326091-326110 + | CCTCCGAA-------------------------------------------------------------------------------------------------------------------------------------------ATTCCTTTAATC | |
| droFic1 | scf7180000451582:1614-1644 - | ATTCCGTA--------------------------------------------------------------------------------------------------------------------------------TGTACCCCGGTGCACCAGTTATA | |
| droEle1 | scf7180000491228:485458-485466 - | TACGGAAAC------------------------------------------------------------------------------------------------------------------------------------------------------ | |
| droRho1 | scf7180000766847:3799-3856 - | CACAGGAATCTTGGGACTTTGGGAATTTGGAGACTTGGCTTCATTTATTTTTT--------------------------------------------------------------A---------------------------------------GCTT | |
| droBia1 | scf7180000302250:74206-74256 + | AATTT-----------------------GGTAGAAGGGTGCCACGCACACCTCTAACTCCCACCATGCGCGAA-------------------------------------------------------------------------------------A | |
| droTak1 | scf7180000415779:155935-155994 - | CAATT------------------------------------------------------------TACGTTAAAATTTGTAAAACAATATTTATGCATTTTAGAAATATATTTAGA---------------------------------------ACTT | |
| droEug1 | scf7180000409538:239137-239138 + | TC------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droSec2 | scaffold_19:646683-646726 - | CACGGAATCCT------TTTCGGAATTTACATATTTCCCTCCACA----------------------------------------------------------------------A---------------------------------------TCTC | |
| droYak3 | 4:904243-904267 - | AATGGAAAAGT------TATAAGAATTTGGA-------------------------------------------------------------------------------------------------------------------------------- | |
| droPer2 | scaffold_87:102155-102195 - | TCCTCAAC------------------------------------------------TTCCCGTCTTGCTCCAACCTTTGCAAAA-------------------------------A---------------------------------------ACTA | |
| droWil2 | scf2_1100000004564:125955-126025 + | CGAAAAAACGT------TTTCGTTATATATTAATAAGTATCCATGGTTTCCAATATCTCCTACAGTACATAA-------------------------------------------A---------------------------------------ATTT | |
| droVir3 | scaffold_13324:2440331-2440378 + | TCGATGTT---------------------------------------------------------------------------------------------------------------TTTGGCTGCGGTTACAATGTACTTGGGGGTTTCGGGAGCT | |
| droMoj3 | scaffold_6500:14920949-14920950 - | TC------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
Generated: 05/15/2015 at 03:24 PM