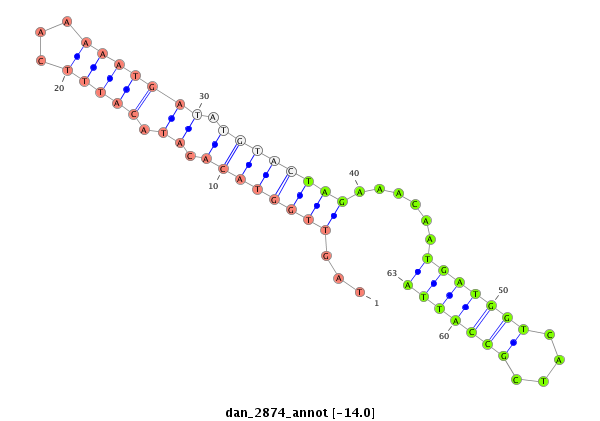
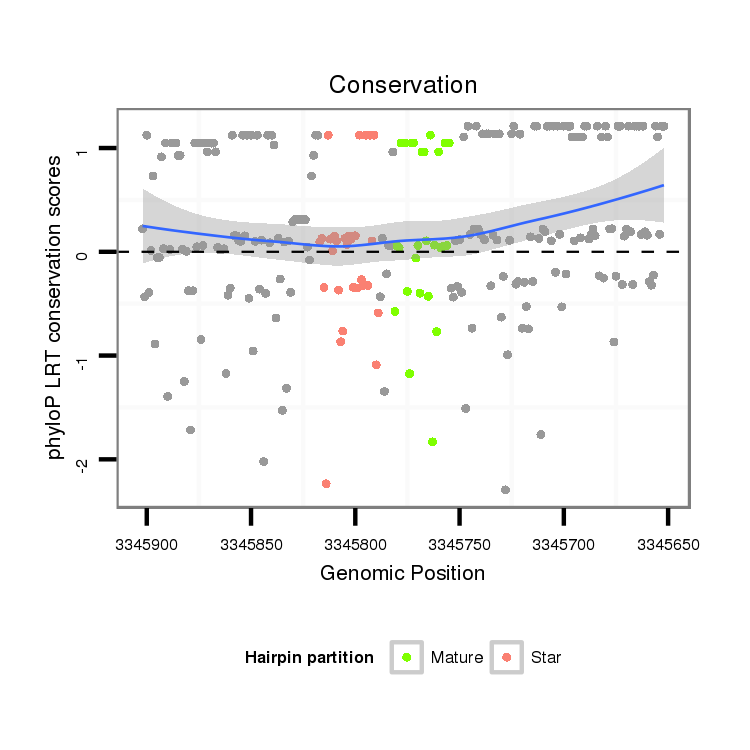
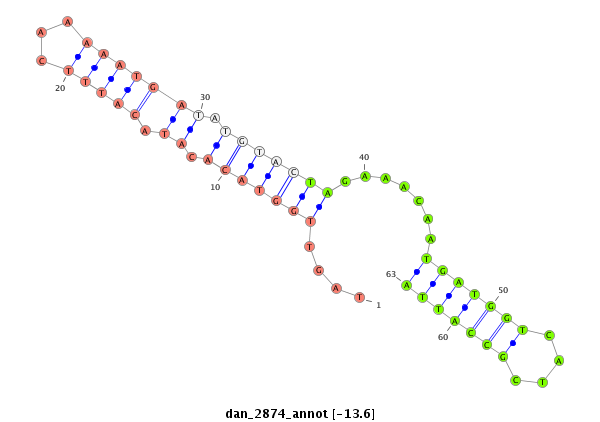
ID:dan_2874 |
Coordinate:scaffold_13340:3345702-3345852 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
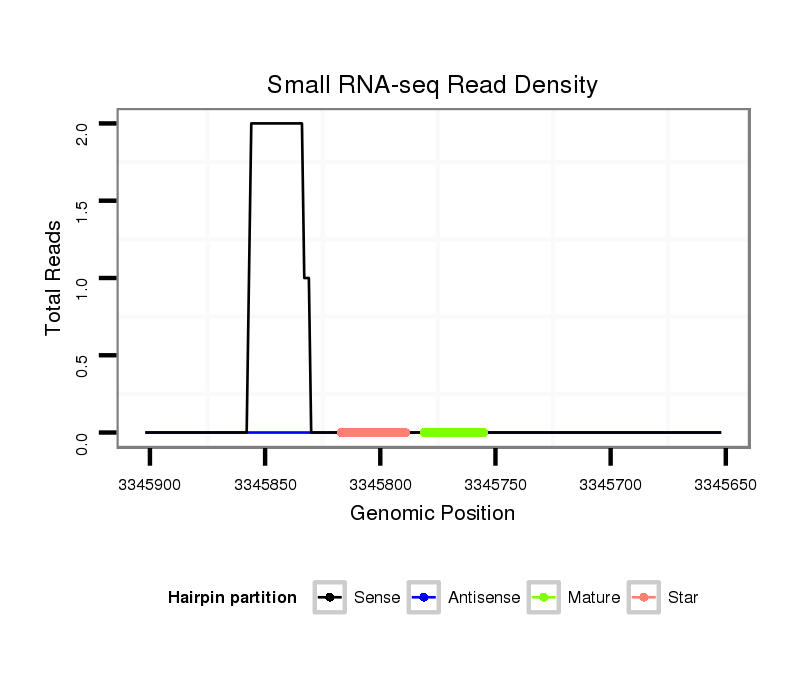
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -13.6 | -13.5 | -13.3 |
 |
 |
 |
exon [dana_GLEANR_18553:5]; CDS [Dana\GF17286-cds]; intron [Dana\GF17286-in]
| Name | Class | Family | Strand |
| Hoana1 | DNA | hAT-hobo | - |
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## AAAAATTAATGTAGAAAATCTGAGATTTCAACTTTGGATGAGTATTCCAAAGATATGGAAACTGCTAGCAACCATTTTTTAATTTTAGTTGGTACACATACATTTCAAAAATGATATGTACTAGAAACAATGATGGTCATCGCCATTACACGGCCTACATAATTCAATATATGGTAAAATTTGATCAATTTCTTTTCTTAGGTGTACCGCGGAAACTGTGGGTGATAGAACCTATGTTTGTTCTGAACTTT *************************************************************************************...((((((((.((.(((((...))))))).))))))))......(((((((....)))))))******************************************************************************************************* |
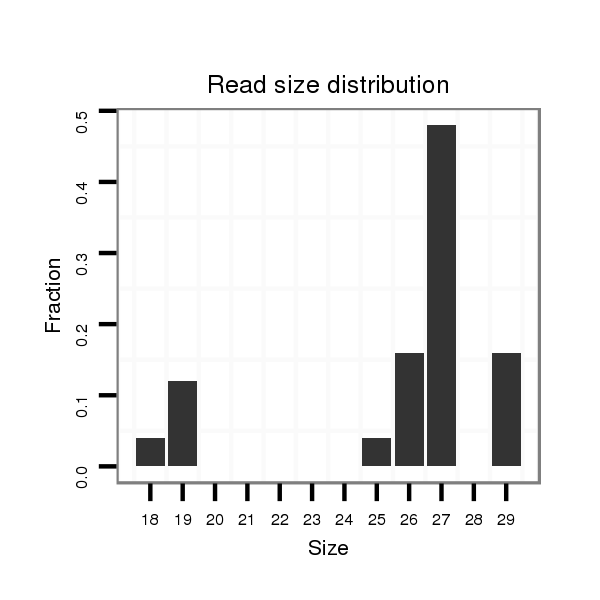
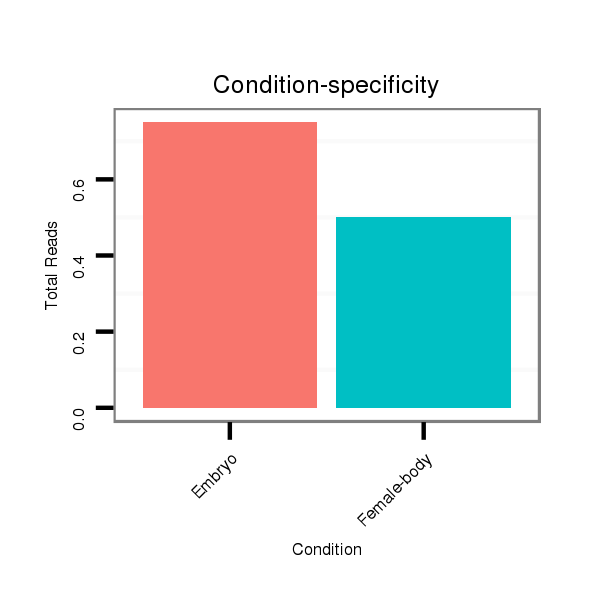
Read size | # Mismatch | Hit Count | Total Norm | Total | M058 embryo |
M044 female body |
V105 male body |
V039 embryo |
|---|---|---|---|---|---|---|---|---|---|
| .............................................TCCAAAGGTATGGAAACTGCTAGCAAC................................................................................................................................................................................... | 27 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .............................................TCCAAAGATATGGAAACTGCTAGC...................................................................................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..............................................CCAAAGATATGGAAACTGCTAGCAAC................................................................................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .........................................................................................................................TAGAAACAATGATGGTCATCGCCATTA....................................................................................................... | 27 | 0 | 20 | 0.55 | 11 | 11 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................TAGGTGTACCGCGGAAACTGT................................ | 21 | 0 | 20 | 0.50 | 10 | 0 | 0 | 10 | 0 |
| .............................AACTTTGGATGAGTATTCCA.......................................................................................................................................................................................................... | 20 | 0 | 20 | 0.25 | 5 | 0 | 0 | 5 | 0 |
| .....................................................................................TAGTTGGTACACATACATTTCAAAAATGA......................................................................................................................................... | 29 | 0 | 20 | 0.20 | 4 | 4 | 0 | 0 | 0 |
| ......................................................................................................................TACTAGAAACAATGATGGTCATCGCC........................................................................................................... | 26 | 0 | 20 | 0.20 | 4 | 0 | 4 | 0 | 0 |
| ......................................................................................................................................GGTCATCGCCATTACACGG.................................................................................................. | 19 | 0 | 20 | 0.15 | 3 | 0 | 3 | 0 | 0 |
| ...................................................................................................................................................................................................TCTTAGGTGTACCGCGGAAACTGTG............................... | 25 | 0 | 20 | 0.15 | 3 | 0 | 0 | 0 | 3 |
| .............................................TCCAAAGATATGGAAACTGCT......................................................................................................................................................................................... | 21 | 0 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 |
| .....................................................................................................................................................................................................................AACTGTGGGTGATAGAACCTA................. | 21 | 0 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 |
| ..........................................................................................................................................................................................................TGTACCGCGGAAACTGTGGGTGATA........................ | 25 | 0 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 |
| ..................................................................................TTTTAGTTGGTACACATACATTTCAAA.............................................................................................................................................. | 27 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| .............................................................................................................................AACAATGATGGTCATCGC............................................................................................................ | 18 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ...................CTGAGATTTCAACTTTGGATGAG................................................................................................................................................................................................................. | 23 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| .......................................................................................................................ACTAGAAACAATGATGGTCATCGCC........................................................................................................... | 25 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ......TAATGTAGAAAATCTGAGATTTCAAC........................................................................................................................................................................................................................... | 26 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ...................CTGAGATTTCAACTTTGGATGAGT................................................................................................................................................................................................................ | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
|
TTTTTAATTACATCTTTTAGACTCTAAAGTTGAAACCTACTCATAAGGTTTCTATACCTTTGACGATCGTTGGTAAAAAATTAAAATCAACCATGTGTATGTAAAGTTTTTACTATACATGATCTTTGTTACTACCAGTAGCGGTAATGTGCCGGATGTATTAAGTTATATACCATTTTAAACTAGTTAAAGAAAAGAATCCACATGGCGCCTTTGACACCCACTATCTTGGATACAAACAAGACTTGAAA
*******************************************************************************************************...((((((((.((.(((((...))))))).))))))))......(((((((....)))))))************************************************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | M044 female body |
M058 embryo |
V106 head |
V055 head |
V105 male body |
|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................................................................................................GGTAATGTGCCGGATGTATTA........................................................................................ | 21 | 0 | 20 | 0.75 | 15 | 14 | 0 | 0 | 1 | 0 |
| ....................................................................................................................................TACCAGTAGCGGTAATGTGCC.................................................................................................. | 21 | 0 | 20 | 0.60 | 12 | 12 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................GGTAATGTGCCGGATGTAT.......................................................................................... | 19 | 0 | 20 | 0.45 | 9 | 9 | 0 | 0 | 0 | 0 |
| ..................AGACTCTAAAGTTGAAACCTACTCAT............................................................................................................................................................................................................... | 26 | 0 | 20 | 0.40 | 8 | 0 | 8 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................GCGCCTTTGACACCCACTATC....................... | 21 | 0 | 20 | 0.25 | 5 | 5 | 0 | 0 | 0 | 0 |
| .....................................................................................ATCAACCATGTGTATGTAAAG................................................................................................................................................. | 21 | 0 | 20 | 0.15 | 3 | 0 | 0 | 3 | 0 | 0 |
| ................................................................................................................CTATACATGATCTTTGTTACT...................................................................................................................... | 21 | 0 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................CCACTATCTTGGATACAAACAAGACTT.... | 27 | 0 | 20 | 0.15 | 3 | 0 | 3 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................TTGACACCCACTATCTTGGATACAAACAA......... | 29 | 0 | 20 | 0.15 | 3 | 0 | 3 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................CCACATGGCGCCTTTGACACCC............................. | 22 | 0 | 20 | 0.15 | 3 | 0 | 3 | 0 | 0 | 0 |
| ......................................................................................................................................CCAGTAGCGGTAATGTGCC.................................................................................................. | 19 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................CCTTTGACACCCACTATCTTGGAT................. | 24 | 0 | 20 | 0.10 | 2 | 1 | 1 | 0 | 0 | 0 |
| ................TTAGACTCTAAAGTTGAAACCTACT.................................................................................................................................................................................................................. | 25 | 0 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 |
| .........................................................................................ACCATGTGTATGTAAAGTTTT............................................................................................................................................. | 21 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 2 |
| ..................................................................................................................................................................................................AAGAATCCACATGGCGCCTTTGACAC............................... | 26 | 0 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 |
| ..................................................................................................................................................................................TAAACTAGTTAAAGAAAAGAATCCAC............................................... | 26 | 0 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................................................TTTAAACTAGTTAAAGAAAAGAATCCAC............................................... | 28 | 0 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................................................TTAAACTAGTTAAAGAAAAGAATCCACC.............................................. | 28 | 1 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................AGCGGTAATGTGCCGGATGTATTAAG...................................................................................... | 26 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................TCCACATGGCGCCTTTGACA................................ | 20 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................................................................................................................................................................CTTGGATACAAACAAGACTTGAA. | 23 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................GTTACTACCAGTAGCGGTAA........................................................................................................ | 20 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................TTTGACACCCACTATCTTGGATA................ | 23 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................GGCGCCTTTGACACCCACTAT........................ | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| .....................................................................................................................................................................................................................TTGACACCCACTATCTTGGATACAAA............ | 26 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................CGCCTTTGACACCCACTATCTTGG................... | 24 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................ATGGCGCCTTTGACACCCACTATCTTG.................... | 27 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................CATGTGTATGTAAAGTTTTTA........................................................................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| .........................................................................................ACCATGTGTATGTAAAGTTTTTACT......................................................................................................................................... | 25 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................ACTACCAGTAGCGGTAATGTGCC.................................................................................................. | 23 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................................................................................................................................................................ACCCACTATCTTGGATACAAACAAGACTT.... | 29 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................CAGTAGCGGTAATGTGCCGGA............................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................GTAGCGGTAATGTGCCGGAT.............................................................................................. | 20 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................ACCAGTAGCGGTAATGTGC................................................................................................... | 19 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droAna3 | scaffold_13340:3345652-3345902 - | dan_2874 | AAAAATTAATGTAGAAAAT--CTGAGATTTCAACTTTGGATGAGTATTCCAAAGATATGGAAACTGCTAGCAA-------CCATTTTTTA-ATTTTAGTTGGTACACATACATTTCAAAAATGATATGTACTAGAAACAATGATGGTCATCGCCATTACACGGCCTACATAATTCAATATATGGTAAAATTTGATCAATTTCTTTTCTTAGGTGTACCGCGGAAACTGTGGGTGATAGAACCTATGTTTGTTCT----GAACTTT |
| droBip1 | scf7180000396730:444189-444434 + | AAAAATAAATGTGTAAAAT---ACACATATAAACTTTGGATGAGTATTCCAAAGATGTGGTAACTGCTAGATA-------TCATTTTTTGCATTTGAGCTGGTATACATACATTTAAAAAATTATATGTACTAAAAACCATGATGGACAACGCAATTACGTGGCCTACATAAATCAATATATGGCAGAA---------TTTATTTTGTTAGGTGTACCGCGGAAACTGTGGGTGATAGAACCTATGTTTGTTTGCTCTCAACATT | |
| droKik1 | scf7180000302692:12459-12708 - | ACAGA--AATGTTGAAAATTTTGGGAGTTTCAACTTTGGCGTCGAATTCCAAAGACATGGGCAATGGTCCACGATTGGGAC-------TA--TTTTTAGTAGTGCCCTGATGTATTGAGAGTAGGTCCTACCGAAAATAATAAAGGTAAGTCCAATTATATAGGCCACGTAATTCAAAATGTCGGCAAACATCGGTTTTTTGTATTCTTAGGTATACCGCGGAACCTGTAGGTGATGGAAAATAATTTTGTATG----GGACTTT | |
| droEle1 | scf7180000490527:7111-7298 - | AAAAT----------------TAGAGATTTTTACTTTGACGTAATATTTAATATATATAGATAATGGTTAAAGCCGGTGAA------------TTTTAATAATGCGATGGTTTATTAAGAGTGTTTG-------------------TCA--------------GCAATACAAT----------GGT--AAATAGGATTTTCATATTCTTAGCTTTACTGCGGAACCCTTAGCTGGAGGAACATAAGTTTATATG----GAACTTT | |
| droRho1 | scf7180000777977:27422-27642 - | GAACT--TGTGTAGAAA-------------------------TCAAAACCAAACAAATTATCAATGGTTGAAGTCAAGAAC-------G----TTTTTTTAACGTGGTGATGATTTGAAAGTGTTTA-ATCAGAAAACGAT-TTGGTCACTTCAATAATATAATCTACGTAATTTAAAATGTCTCCAAACTTAAAATTTTCTTATTCTAAACTATACTGCCGAACTTGTGGTTGATAAAACATAAGTTTGTATG----GGACTTT | |
| droBia1 | scf7180000302087:8787-8888 - | A-------------------------------------------------------------------------------------------------------------------------------------------------------------CGTAGGCAACATAGTTCAATGTGTGAAAAAACATCG--TATTTGCATTCTCAGCTATACCGCGGAACCTGTAGGTGATTAAACACAAGTTTGTATA----AGACTTT | |
| droTak1 | scf7180000415242:413291-413539 + | ACAAAT----TTTGAAAATTTTGGGAGTTTCAACTTTGGCGTCGAATTCCAAAGACATGGGCAATGCGTCACGATTGAGAC-------TA-ATTTTTAGTAGTGCCGTGATGTATTGAGAGTAGGTCCTACCGAAAATAATAAAGGTAAGTCCACTTATATTGGCCACGTAATTCAAAATGTCGGAAAACATCGGTTTTTTGTATTCTTAGGTATACCGCGGAACCTGTAGGTGATGGAAAATAATTTTGTATG----GGACTTT |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
Generated: 05/18/2015 at 05:49 AM