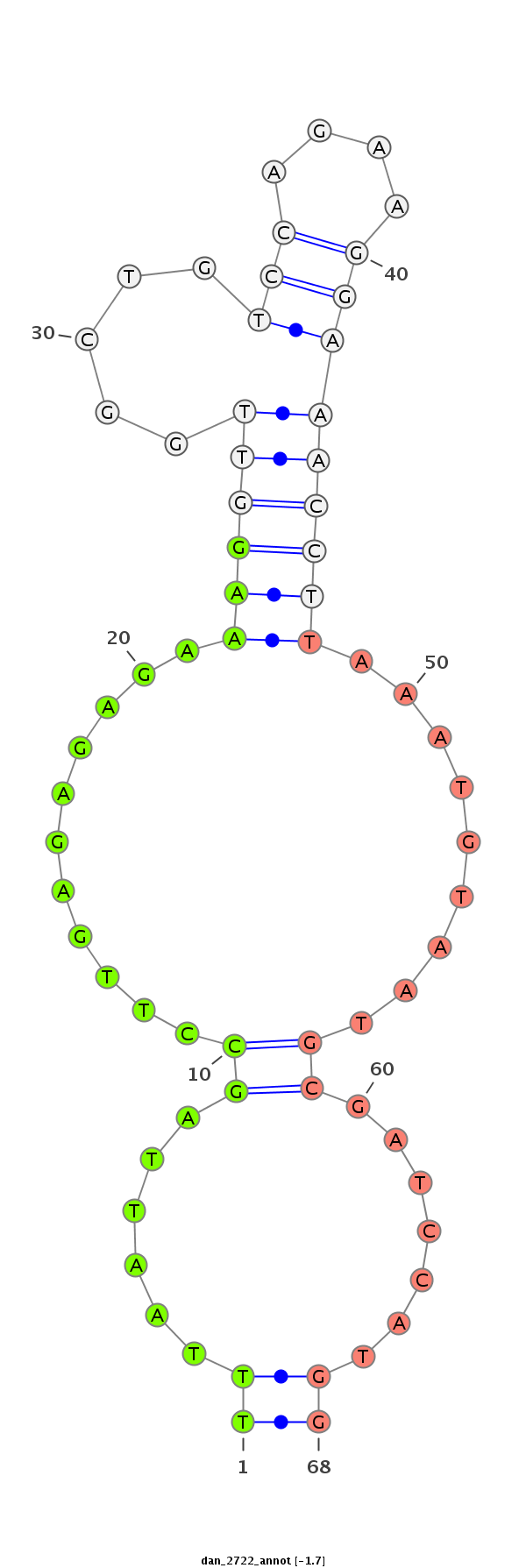
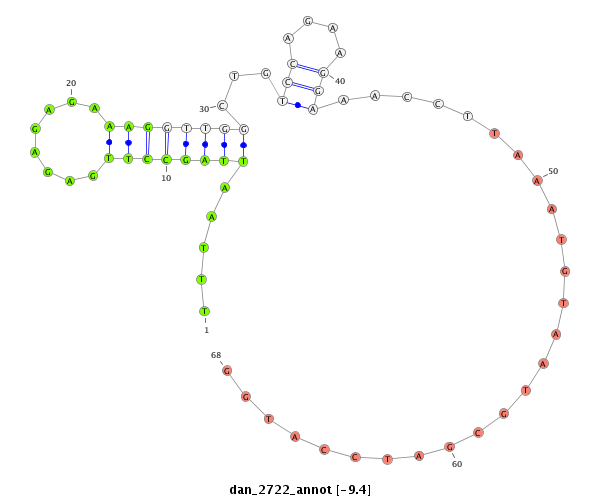
ID:dan_2722 |
Coordinate:scaffold_13337:19347974-19348124 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
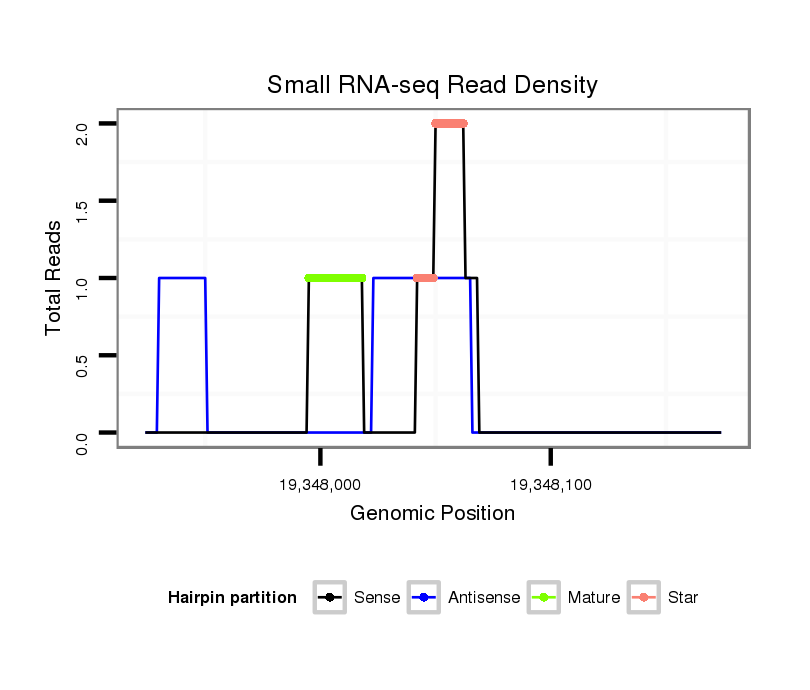
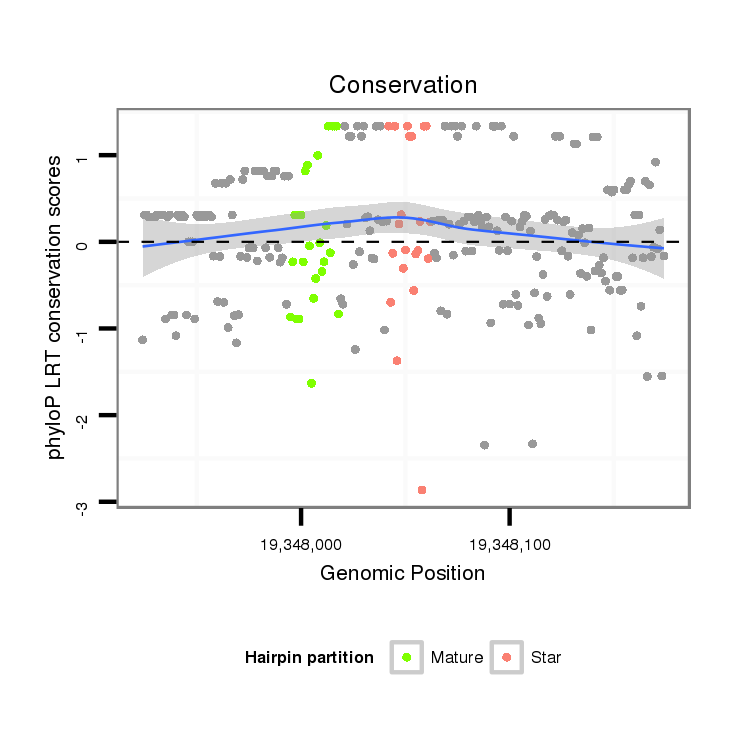
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -9.4 | -9.0 | -8.8 | -8.8 |
 |
 |
 |
 |
exon [dana_GLEANR_10796:3]; CDS [Dana\GF10838-cds]; intron [Dana\GF10838-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GAACCCGAAATAGAGCATGAGGAGCATGAAGAACCGATCCCAAAACCAAAGTATTATAAACGCAAGAAGTCTTTAATTAGCCTTGAGAGAGAAAGGTTGGCTGTCCAGAAGGAAACCTTAAATGTAATGCGATCCATGGTTAGGGAAATGTCAAACTTTCATGCCTCGTTTTTAATTACATACAAAAAACAAAAAAAACGCTCCACATAATTAAGTTAAAACCTCGAATCCTACATTCAATACCTGATTGA ***********************************************************************((......((...........((((((.....(((....))))))))).........)).......))**************************************************************************************************************** |
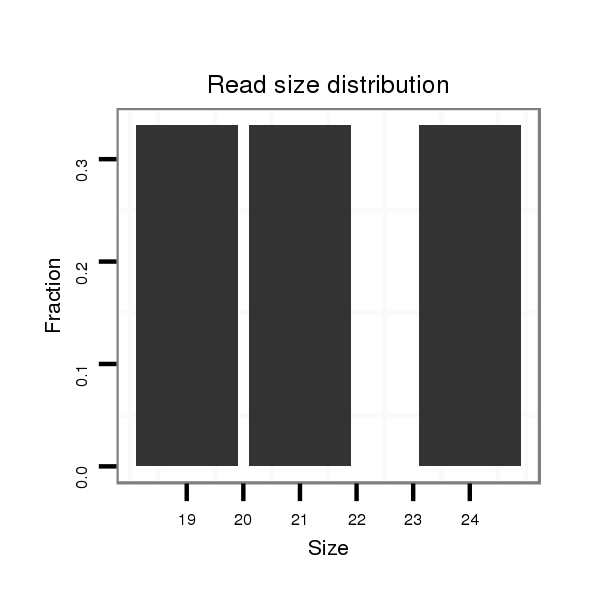
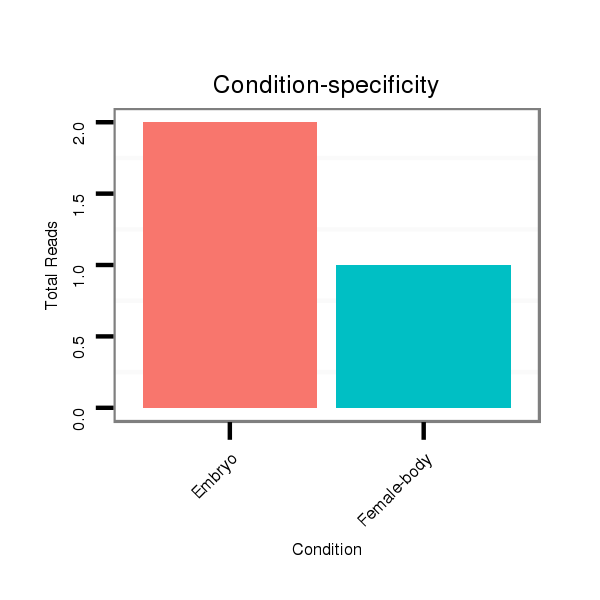
Read size | # Mismatch | Hit Count | Total Norm | Total | M058 embryo |
V039 embryo |
M044 female body |
V055 head |
V105 male body |
|---|---|---|---|---|---|---|---|---|---|---|
| .......................................................................TTTAATTAGCCTTGAGAGAGAAAG............................................................................................................................................................ | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................TAAATGTAATGCGATCCATGG................................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................ATGCGATCCATGGTTAGGG.......................................................................................................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................................................................................................................AAAAAAAACGCTCCGCAGAA......................................... | 20 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................GAGGAGCCTGAATATCCGATC.................................................................................................................................................................................................................... | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 |
| ......................TGGATGAAGAACGGATCCCAA................................................................................................................................................................................................................ | 21 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................................................................................................................AAAGACAGAAAAAACGCACCAC............................................. | 22 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................AAAAAAACGCTCCACACCA......................................... | 19 | 2 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................AAAACAAAAAAAACTCCCCACA............................................ | 22 | 2 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................AAAAACAAAAACAACCCTCCCCA............................................ | 23 | 3 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................CAGAAGGATAACTTAAATGC.............................................................................................................................. | 20 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 0 | 1 |
| ........................................................................................................GCAGAAGGATAACTTAAATG............................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| ..........................................................................................................................................................................................AAACACAAAAAACGCACCAGA............................................ | 21 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................AAGTTAGAGGCTCGAATCC.................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................AAATGTCGAACTTTCTTG........................................................................................ | 18 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................GAGGAGCATGGATATCCGA...................................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..........................................................................................................................................................................................AACCAAAAAAAACGCACCA.............................................. | 19 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................AAACAAAAAAAACGCTAGA.............................................. | 19 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
|
CTTGGGCTTTATCTCGTACTCCTCGTACTTCTTGGCTAGGGTTTTGGTTTCATAATATTTGCGTTCTTCAGAAATTAATCGGAACTCTCTCTTTCCAACCGACAGGTCTTCCTTTGGAATTTACATTACGCTAGGTACCAATCCCTTTACAGTTTGAAAGTACGGAGCAAAAATTAATGTATGTTTTTTGTTTTTTTTGCGAGGTGTATTAATTCAATTTTGGAGCTTAGGATGTAAGTTATGGACTAACT
****************************************************************************************************************((......((...........((((((.....(((....))))))))).........)).......))*********************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M044 female body |
M058 embryo |
V039 embryo |
V055 head |
|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................ACAGTTTGGAAGTACGGAGC................................................................................... | 20 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ........TTATCTAGTACTCCTCGTACTTCTTGGC....................................................................................................................................................................................................................... | 28 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ...................................................................................................CGACAGGTCTTCCTTTGGAAT................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ........................................................................................................................TTACATTACGCTAGGTACCAAT............................................................................................................. | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ......CTTTATCTCGTACTCCTCGTA................................................................................................................................................................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................AGGATGTGAGTTATGGAATAT.. | 21 | 3 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 |
| .......................................................................................................................................................GTTTGAGAGTAGGGAGC................................................................................... | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droAna3 | scaffold_13337:19347924-19348174 + | dan_2722 | GAACCCGAAATAGAGCATGAGGAGCATGAAGA------ACCGATCCCAAAACCAAAGTATTATAAACGCAAGAAGTCTTTAATTAGCCT-TGAGAGAGAAAGGTTGGCTGTCCAGAAGGAAACCTTAAATGTAATGCGATCCATGGTTAGGGAAATGTCAAACTTTCATGCCTCGTTTTTAATTACATACAAAA---AACAAAAAAAACGCTCCACATAATTAAGTTAAAACCTCGAATCCTACATTCAATACCTGAT------------------------------------------------------------------------------TGA |
| droBip1 | scf7180000396360:541066-541312 + | GAACCCGAAATTGAATCTGAGA---TTGAAGAATTACAACCGGTCTCCAACGAAAAGTATTTTAAACGCAAGCGGTCGATTTTAAGAAT-TGAGCAAGAAAAAATAGCTGTCCAGAAGCAGACCTTAAATATAATGCGGTCGATGGTTAAGGAAATTTCAAACTTTCATGCGTCATTTTTAATTACTTACAAGACACAAAACAAAAAACGCTCTACGTAGTTAAATTCAGACC----------CATTAATTAGCTGAT------------------------------------------------------------------------------GGA | |
| droKik1 | scf7180000302686:355171-355337 - | G-------------------------------------------------------------------------------------------AGGAAAAAAAACTGGCTATTCAAAAAGAAATACTCCACGTAATGCAAACAATGGCAAACGATCTTTCCAGTACATATACCACCTTTCTAGATGCTTACAAAACAAGTCCGACGAAACGCTTTTGATAGTTCAAATTAGATGTAGATAAATACAA---AAAACCAAA------------------------------------------------------------------------------TAT | |
| droFic1 | scf7180000454113:2398535-2398678 + | A------------------------------------------------------------------------------------GCTGTGGAAGAAAAAACTTTGGCCGTTCAACGAAAAACCCTTAAAGCCATGCAAGCAATGGCCGAAGATCTTTCCAGTTTTCATGAAAATTTCCTTAAGGCGATCAAACCAACATTCCAAAAACACGCTTAATAGCC---------------------------------AAT------------------------------------------------------------------------------TAA | |
| droEle1 | scf7180000491193:4193002-4193212 - | A------------------------------------------------------AGCGCTATAAGCGCAAACGGACCA----AAGCTGTGGGAGAAGAAATGTTGGCCTTGCAAAGAGAAACACTCGACGTCCTGCGAAATATTGCAAAAGACCTTTCCAGTTTTCATGAAAAGTTCTTAAATGCACTCAAGCCCATGCATCAGCGACGCTCTTCATAGTTATATTAAGT---------------------------TATTTAAGGAATACTATTAGTAGTTATGA--------C---------------------------TTTC--------TTTT | |
| droRho1 | scf7180000773422:10816-11071 + | C---------------------------------------TGAACTCAA-CGCTAAACGCTATAAGCACAAACGGGCCA----AAGCAATGGGAGAAGAAATGTTGGCTCTTCAAAGAGAAACTCTCAACGTCATGCGAACGATGGCAAAGGACCTTTCCAGCTTTCATGACAAGTTCTTGAAAGCACTAAAACCAACTCACCACAAACGCTCTTCATAGCTAGAATAAAT---------------------------TACTTAAAAAATTATATTTGTTATTATGATATGACAACATTCTTTCAA----AAGGTGCAAAATATTTTTG--------CT | |
| droBia1 | scf7180000302428:1388803-1389008 + | ------------------------------------------------------------------------------------------------AGAAATGTTGGCCCTTCAAAGGGAAACTCTCAAGGTCTTGCAGACAATTTCAGAGAACCTCTCCAGTTTCCACGATAAGTTCTTGAGAGCACTCAAGCCAAGACGCGAGAAGCGCCCTA--------GACTAAGT---------------------------TAGCTAAGAGCT-GCATTTGTTAT-ATATTAAGAGAATTTTCCTACATACGTAATATGCAAAATATTTTTGCAAAACTTGT | |
| droTak1 | scf7180000415420:174209-174345 + | T-----------------------------------------------------------------------------------------TGGCGAAGAAATGCTGGCCCTTCAAAGGGAAACTCTAAATGTTATGCGAACTATTGCAGAGGATCTTTCCAGTTTTCATGACAAGTTCTTGAAAGCATTCGCGCCAAAACGCCAGAAACGCTCTA--------GACTAAGC---------------------------TAA------------------------------------------------------------------------------ |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
Generated: 05/18/2015 at 05:13 AM