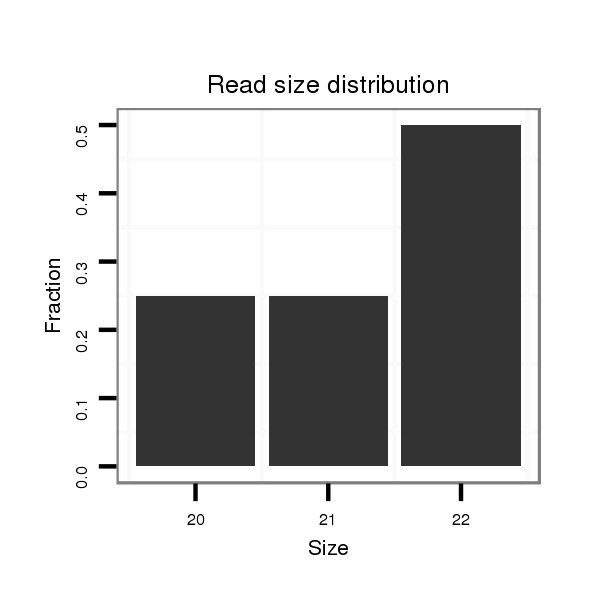
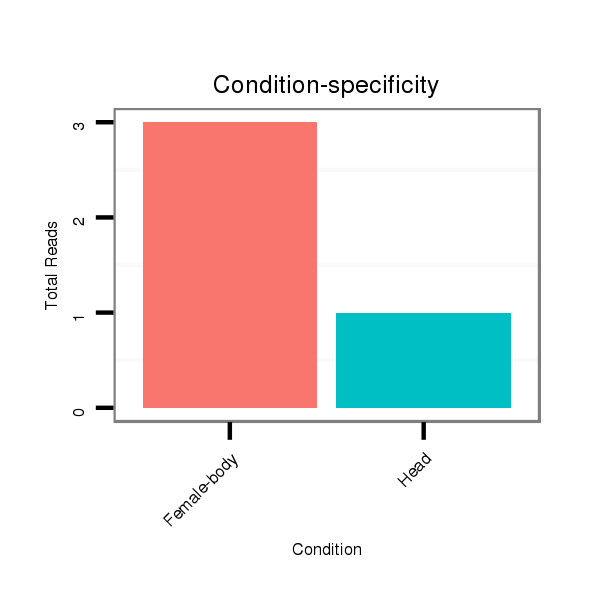
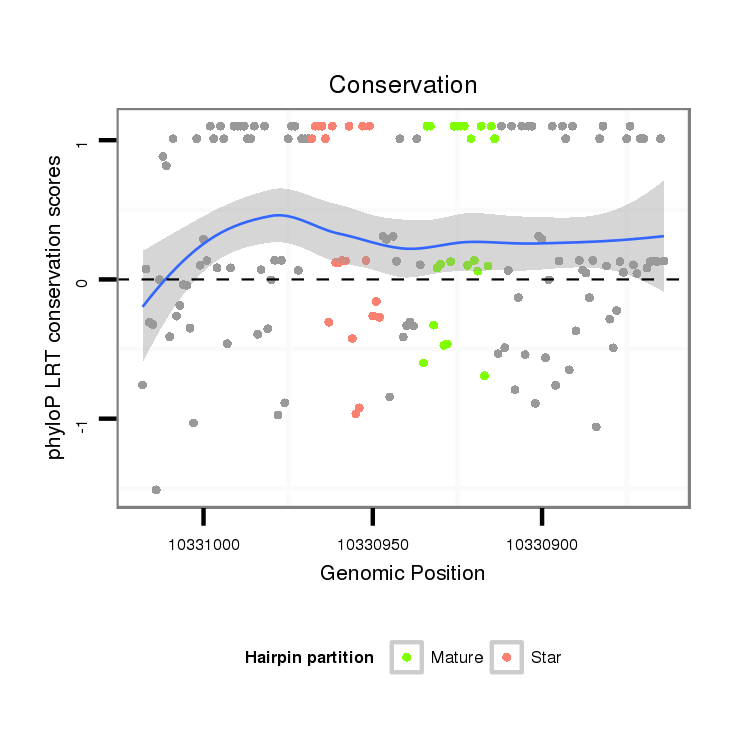
| .....................................................................................ATGACTCAATTCCTTTACAGT................................................. |
21 |
0 |
1 |
20.00 |
20 |
12 |
4 |
3 |
1 |
0 |
| .....................................................................................ATGACTCAATTCCTTTACAGTT................................................ |
22 |
0 |
1 |
2.00 |
2 |
1 |
1 |
0 |
0 |
0 |
| ...................................................................................AAATGACTCAATTCCTTTACAG.................................................. |
22 |
0 |
1 |
2.00 |
2 |
0 |
2 |
0 |
0 |
0 |
| ......................................................................................TGACTCAATTCCTTTACAGT................................................. |
20 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
| ...................................................................................AAATGACTCAATTCCTTTACAGT................................................. |
23 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
| .....................................................................................ATTACTCAATTCCTTTACAGT................................................. |
21 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................................................GTAAGGATAATTGATAAATTC.................................................................................... |
21 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .....................................................................................ATGACTCAATTCCTTTACAG.................................................. |
20 |
0 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
| ........................................................................................................................GTGATCCTGTGTCTGCAGCCAG............. |
22 |
3 |
2 |
0.50 |
1 |
0 |
0 |
0 |
1 |
0 |
| ......................................................................................................................................ACACCCAGCGAACGGCCTCG. |
20 |
3 |
3 |
0.33 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................CAAACACACAATTGTAGTTAGT................................................................................................................... |
22 |
3 |
3 |
0.33 |
1 |
0 |
0 |
0 |
0 |
1 |
| ...............................GGAGATACTTTTGAAGGCGGT....................................................................................................... |
21 |
3 |
6 |
0.17 |
1 |
0 |
0 |
0 |
1 |
0 |
| ................................................................................................................ATTTATGGGTGATCCTGT......................... |
18 |
2 |
8 |
0.13 |
1 |
0 |
0 |
0 |
1 |
0 |
| ......AGCTAGAGGAGCCAAGCA................................................................................................................................... |
18 |
2 |
11 |
0.09 |
1 |
0 |
0 |
0 |
1 |
0 |
| .....................................................................................................................................TGAACACAGCCGGCGGCCTC.. |
20 |
3 |
12 |
0.08 |
1 |
0 |
0 |
0 |
1 |
0 |
| .................................................................................................................ATTATGGATGATCCTGCATGT..................... |
21 |
3 |
16 |
0.06 |
1 |
0 |
0 |
0 |
1 |
0 |
| ......................CACACCATTGGAGTTGGTTT................................................................................................................. |
20 |
3 |
20 |
0.05 |
1 |
0 |
0 |
1 |
0 |
0 |
| ....................................................................................................................ATGGGAGATCCGGTATCGG.................... |
19 |
3 |
20 |
0.05 |
1 |
0 |
0 |
0 |
1 |
0 |