
ID:dan_251 |
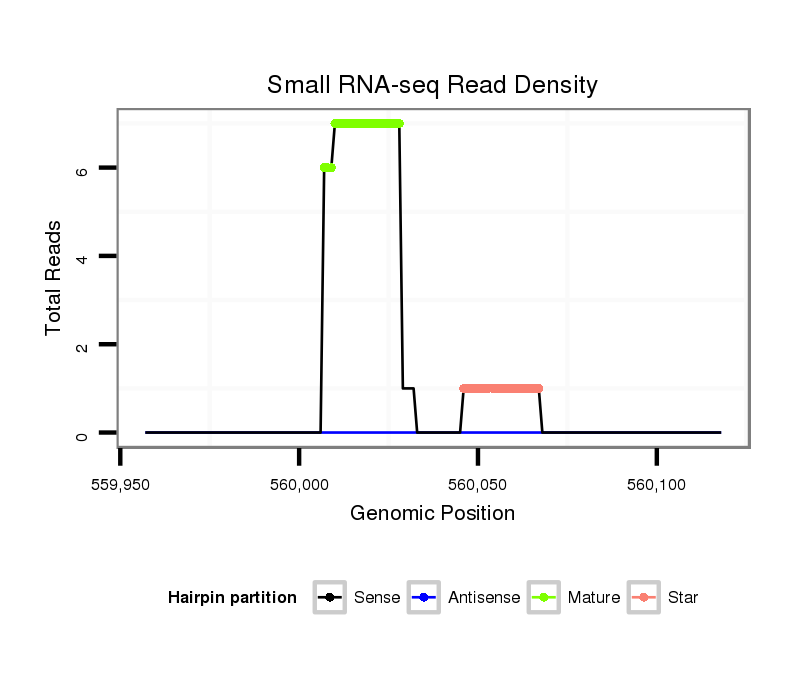
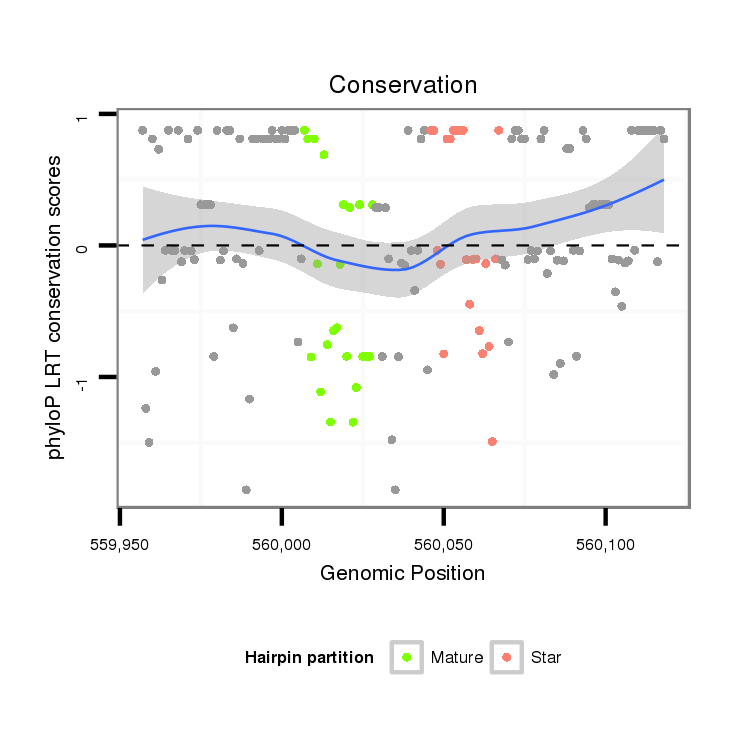
Coordinate:scaffold_12903:560007-560068 + |
Confidence:candidate |
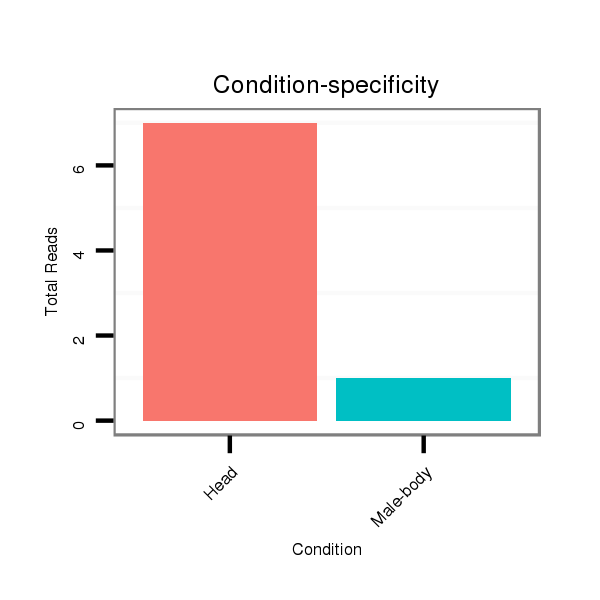
Class:Testes-restricted |
Genomic Locale:intergenic |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -31.2 | -31.1 | -31.1 |
 |
 |
 |
intergenic
No Repeatable elements found
|
AAGCGAAAAAAAAACCGAAAAATTCCTTATCGAGGGAGGCAGGTCTTTGGTGGCTACTTAAATTGCTTGTGTGGCGAGAGAGTACACACATAAGCGATTAGAGTAACCTTTCGGGATCCGTTCCAGCAACAGCAACTGCATATATAGAGTATATTATTTATG
***********************************..(((.((((((....((.(((((.(((((((((((((.............))))))))))))).))))).))....)))))).)))....************************************ |
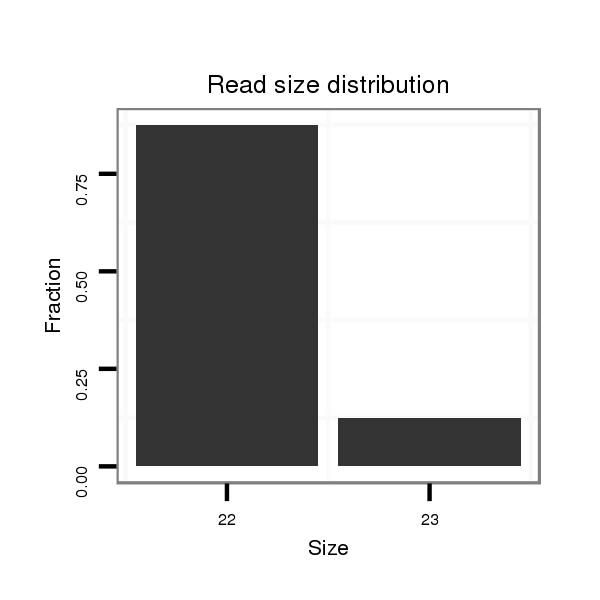
Read size | # Mismatch | Hit Count | Total Norm | Total | V106 head |
V055 head |
M044 female body |
V105 male body |
|---|---|---|---|---|---|---|---|---|---|
| ..................................................TGGCTACTTAAATTGCTTGTGTGT........................................................................................ | 24 | 1 | 1 | 7.00 | 7 | 3 | 0 | 4 | 0 |
| ..................................................TGGCTACTTAAATTGCTTGTGT.......................................................................................... | 22 | 0 | 1 | 6.00 | 6 | 2 | 3 | 0 | 1 |
| ..................................................TGGCTACTTAAATTGCTTGTGTGTT....................................................................................... | 25 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ..................................................TGGCTACTTAAATTGCTTGTGTT......................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .....................................................CTACTTAAATTGCTTGTGTGGCT...................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ...................................................GGCTACTTAAATTGCTTGTGTGT........................................................................................ | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 |
| .........................................................................................ATAAGCGATTAGAGTAACCTTT................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ..................................................TGGCTACTTAACTTGCTTGT............................................................................................ | 20 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| .....................................................CTACTTAAATTGCTTGTGTGGCG...................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .............................TCGGGGGAGGCAGATCT.................................................................................................................... | 17 | 2 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 |
| ..................................................................................TACAAACATAAGCGCTTACA............................................................ | 20 | 3 | 14 | 0.07 | 1 | 0 | 1 | 0 | 0 |
|
TTCGCTTTTTTTTTGGCTTTTTAAGGAATAGCTCCCTCCGTCCAGAAACCACCGATGAATTTAACGAACACACCGCTCTCTCATGTGTGTATTCGCTAATCTCATTGGAAAGCCCTAGGCAAGGTCGTTGTCGTTGACGTATATATCTCATATAATAAATAC
************************************..(((.((((((....((.(((((.(((((((((((((.............))))))))))))).))))).))....)))))).)))....*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M058 embryo |
|---|---|---|---|---|---|---|
| ................................................CCACCGATGAATTTAAGTAA.............................................................................................. | 20 | 2 | 2 | 0.50 | 1 | 1 |
| ..CGCTTTTTATTTGGCTTTT............................................................................................................................................. | 19 | 1 | 5 | 0.20 | 1 | 1 |
| Species | Coordinate | ID | Type | Alignment |
|---|---|---|---|---|
| droAna3 | scaffold_12903:559957-560118 + | dan_251 | candidate | AAG-CGAA---------AAAAAAACCGAAAAATTCCTTATCGAGGGAGGCAGGTCT-TTGGTGGCTACTTAAATTGCTTGTGTGGCGAGAGAGTACACACATAAGCGATTAGAGTAACCTTTCGGGATCCGTTCCAGCAAC-------------AGCAACTGCATATATAGAGTATATTATTTATG |
| droBip1 | scf7180000395925:48291-48454 - | AGTGCAAAAAACTGACAAAAAAAACCGAAAAACTTCTTATCTTCGGAGGCAGGTCT-TTAGTGTCCTCACTATTCGGGTACATGGTGATTTTATACACATATACACGATTAGCGTTGACGTTTAAGATCCGTTCCAGC-------------------ACCTGCATATATA--TAGTATTATTTATG | ||
| droEle1 | scf7180000490751:845095-845240 - | AAA-CA-C---------TAGGAAGCTTA-----TCTTTGGCGGTGGGGGCAGGTCTCTTTGTGCCT---AGGT--------------CTGAAATGGTCACATTCCCGATTATATGGCCAAGTCATGATCCTCGTCACTGGTACTAAAAAGAAATTGCTGTTG-------CCGTTGTTTTATTTATG | ||
| droTak1 | scf7180000415843:938913-939061 + | ATT-CGAA---------TAGGAGGCTTA-----TCTTTTGCGCTGGGGGCAGGTCT-TTTATGCCTCCTCGTT--------------CCCAAATGCTCATATTCCCGATTATATGGCCGTGTCATGATCCTCGTCACTTGAACGAGAAAGAAACTGCTGTTG-------CCGTTGTTTTATTTGTG |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
Generated: 10/20/2015 at 07:29 PM