
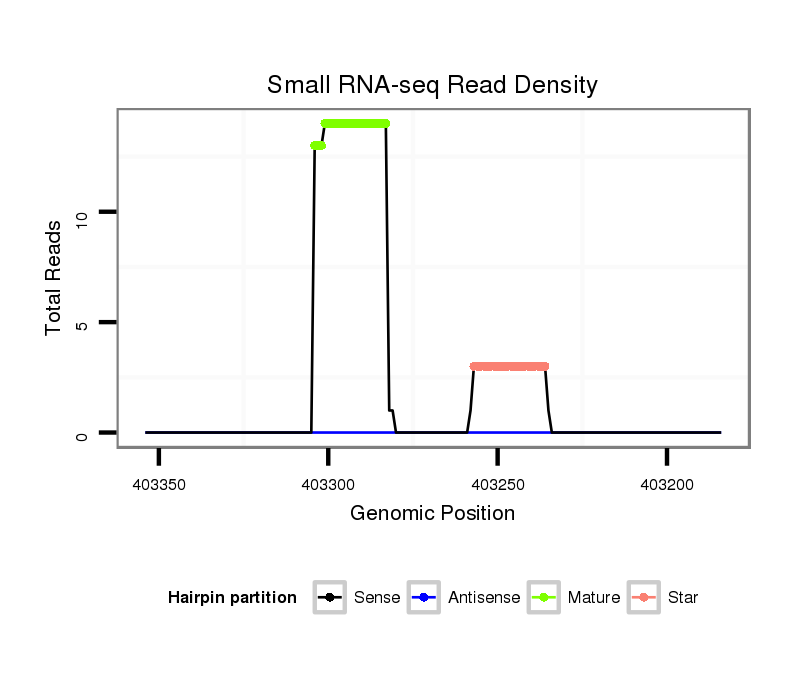
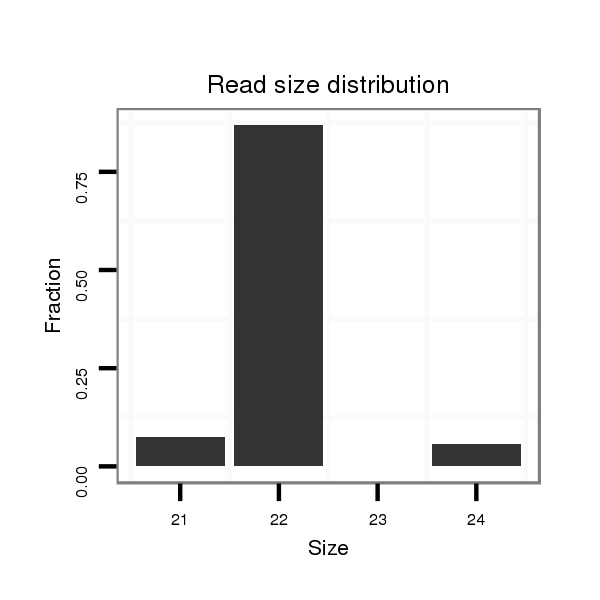
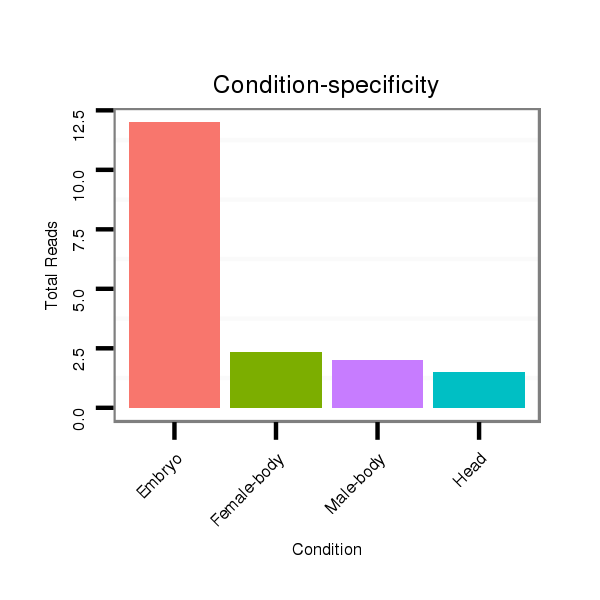
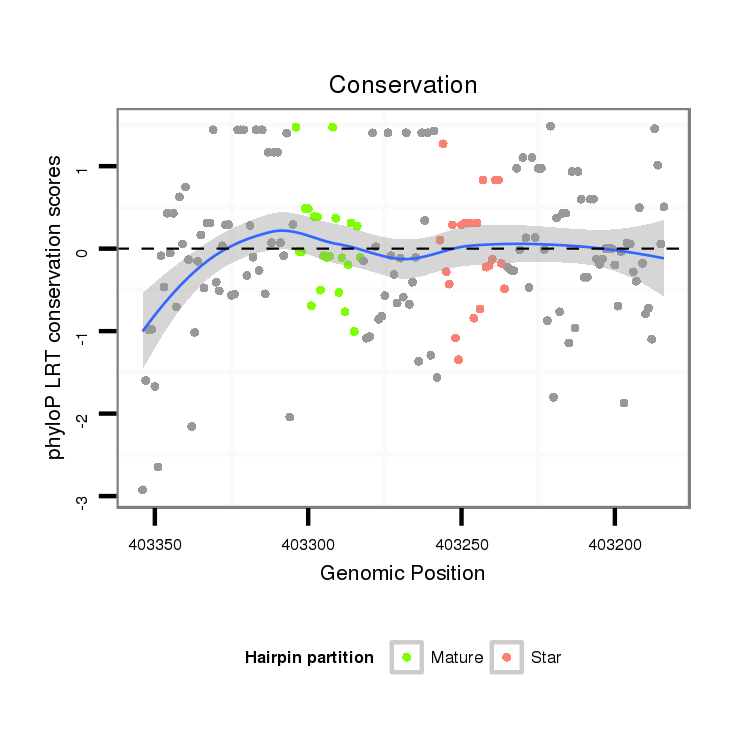
ID:dan_245 |
Coordinate:scaffold_13137:403234-403304 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -33.6 |
 |
intergenic
| Name | Class | Family | Strand |
| AT_rich | Low_complexity | Low_complexity | + |
| mature | star |
|
GCATTTAATTTATTTAATTTTAAATAAAAAGTTTTTTTGTTTGTACATCAAAAATATTCTTATTCGACACGTGTAACAATAACAAAAATATAAACACGTGCCGGGTAAGAATATTTTGAATGTATAATTTTTTTACATATATTACATATATATTATTATTCGTCGCCAAAA
***********************************.....((((((((..((((((((((((((((.(((((((...................))))))).))))))))))))))))..)))))))).......************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | M058 embryo |
V106 head |
M044 female body |
V105 male body |
V039 embryo |
|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................AAAATATTCTTATTCGACACGT................................................................................................... | 22 | 0 | 2 | 13.50 | 27 | 20 | 1 | 0 | 4 | 2 |
| .................................................................................................GGGCCGGGTAAGAATATTTTGAAA.................................................. | 24 | 2 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 |
| .................................................................................................GTGCCGGGTAAGAATATTTTGA.................................................... | 22 | 0 | 1 | 2.00 | 2 | 0 | 1 | 1 | 0 | 0 |
| .................................................................................................GTGCCGGGTAGGAATATTTTG..................................................... | 21 | 1 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 |
| .................................................................................................GTGCCGGGGAAGAATATTTTGA.................................................... | 22 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ................................................................................................CGTGCCGGGTAAGAATATTTTGAA................................................... | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................................................ATATTCTTATTCGACACGTGT................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..................................................AAAATATTCTTATTCGACACGC................................................................................................... | 22 | 1 | 3 | 0.67 | 2 | 2 | 0 | 0 | 0 | 0 |
| ................................................................................................CGGGCCGGGTATGGATATTTTG..................................................... | 22 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 |
| ..................................................AAAATATTCTTATTCGACACG.................................................................................................... | 21 | 0 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..................................................ATAATATTCTTATTCGACACGT................................................................................................... | 22 | 1 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................CGTGCCGGGTAAGAATATTCTGT.................................................... | 23 | 2 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................AATATAAACACGTGCCGT................................................................... | 18 | 1 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 1 |
| ............................................................................................................................................ATTACCTCTATATTATTAT............ | 19 | 2 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
|
CGTAAATTAAATAAATTAAAATTTATTTTTCAAAAAAACAAACATGTAGTTTTTATAAGAATAAGCTGTGCACATTGTTATTGTTTTTATATTTGTGCACGGCCCATTCTTATAAAACTTACATATTAAAAAAATGTATATAATGTATATATAATAATAAGCAGCGGTTTT
*************************************.....((((((((..((((((((((((((((.(((((((...................))))))).))))))))))))))))..)))))))).......*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V106 head |
|---|---|---|---|---|---|---|
| ...............................AAAAAAATGAACATGTAG.......................................................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droAna3 | scaffold_13137:403184-403354 - | dan_245 | GCATTTAATTTATTTA-----ATTTTAAATAAAAAGTTTTTTTGTTTGTACA------------------------------------------TCA--AAAATATTCTTATTCG--------------------------------------------------------------------------------ACACGTGTAACAATAACAA----AAATATAAA--CA-------CGTGCCGGGTAAGA-----------ATATTTTGAATGTATAATTTTTTTA-------------------CATATATTACATATATATTATTATTCGTCGCCAAAA |
| droBip1 | scf7180000394722:28262-28428 + | AAATTCGATTTTTCTA-----C---------------------------------TTCCCAAATGAAAAA--GACTTCTTC-----TTCTGA---AA--AAAATATTCTTATTTG--------------------------------------------------------------------------------GGACGTGCCACTAAAACAA----TATAACAAAGAAA-------TGTGCCTCGTAAAAAAATGATGGATG----------------------TGCTTTTTAT-GATTTTTATACCTACAAT------------TTTTTTGCTACTAAAA | |
| droKik1 | scf7180000302469:477473-477630 - | TCAAAAATTTTAACAA-----A---------------------------------TTTTTAACTACAAAA--AATTTTATA-----TTTTTAAGTTAGTAAAATATTTTTATATT--------------------------------------------------------------------------------AAATATGGAAAAATATTTA----AACTAAAAA--AA-------A---------------ATTATGAAAATATATTAAACTTATAATTTTATTA-------------------TATAAAATATGTAAAA------ATATTT-------A | |
| droFic1 | scf7180000454042:92437-92575 + | CTTCCCAAAACATTTA-----TG--------------------------------CTTG--AAAAAATATAAGATT--------------------------TT-----------------------------------------------------------------GTTAATATTCTAAATACCGTTATCCGCAGAAATTAACGCTAATAG----TAAAGTAAA--TA-------TATTG----------------------------------TGTTT---------TTAAA---AGTAAAAATTTT---------------AAACATTTTTATAAATA | |
| droEle1 | scf7180000491349:1154091-1154256 + | GTGTCTGGTATTTTTT-----G---------------------------------TTTA--AGAATATGAAAGTGTTTTACAATTATTTTTGA-TTT--AGAATATTCTTATTTG--------------------------------------------------------------------------------AAAGATATCACAGCAACGG----AAAAACAAA--A---------------------------------ATTCATTGCTTTTTTATTTTTTCTCCGATTAATACAAAAATATATACA---------------AAATATTGTTGAAAAAA | |
| droRho1 | scf7180000764289:8406-8559 - | AATTAA--------TATCTGGATATCAAATTAAATGTTTCGTTTTATATATTTAAATT----AA-------------T----------------TAT--AAAAACATACCTTTTAACACAAGACAAATAAAACTGCTTGACTAAATAAGAATAGATTTAAATGCAGATTGATAATATTAGATATGAT-------------------------------------------------------------------------------------------------------------------------------------------GTTATTCGT-------A | |
| droBia1 | scf7180000301916:7261-7379 + | AAAAACAATTTTTTTA-----A---------------------------------TATATAAAAACATAAAAATATT-------------------------------------------------------------ATGAAAATTAAAACAAAAACACATACAAACAAAT----------------------------------------------------------AAATAATACAT----------------------------------TATAATATTTATA-------------------TATATATT------------ATATATATTATTA--T | |
| droTak1 | scf7180000414528:11362-11510 - | CAATTA--------TTACAGTTGATTTTACC-AAGTTTTTTTTGTGTGTACT-------------------------T----------------TCT--AATATTTTCTCGTTCGGCACGGTGCAATAGAAAATGCCTG--TGAATGAGAAAATATTAGAAA-----------------------------------------------------------------------------------------------------G----------------------TCTATTTAAT-GACGAGTGTA-------------ATA---ATTTTTTGTCACAAAAT | |
| droWil2 | scf2_1100000004963:459184-459322 + | TTTA--------TTTAGTTTGAAAAAAAAATTATAATTTCTATGTTTGTACA------------------------------------------TCA--ATTATGTATGTATTCG--------------------------------------------------------------------------------AAATAAGAAA--TTATTGGAAAAAAAAGTAGA--TA-------AAT----------------------G----------------------TCCAATTTACACATGTATGTATGTA---------------CATATTCGTTCCCA--A | |
| droVir3 | scaffold_12855:9978064-9978104 + | TTGCTTATTTTATTTT-----TTTATAAATTTTAAATTTTTCTATT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
Generated: 05/15/2015 at 03:08 PM