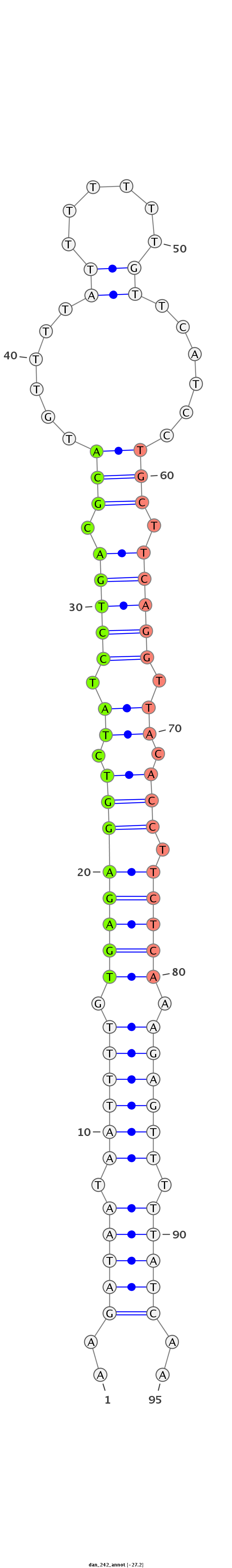
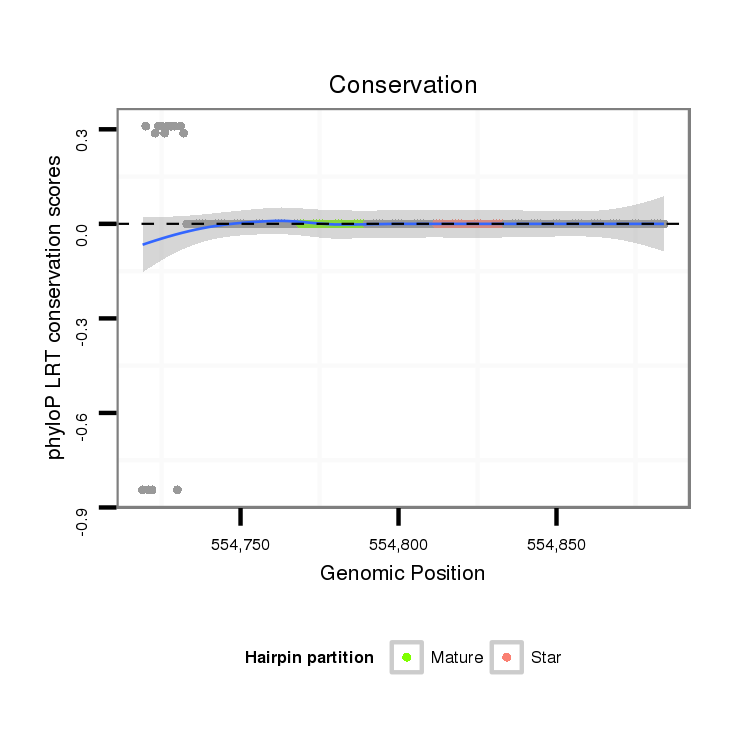
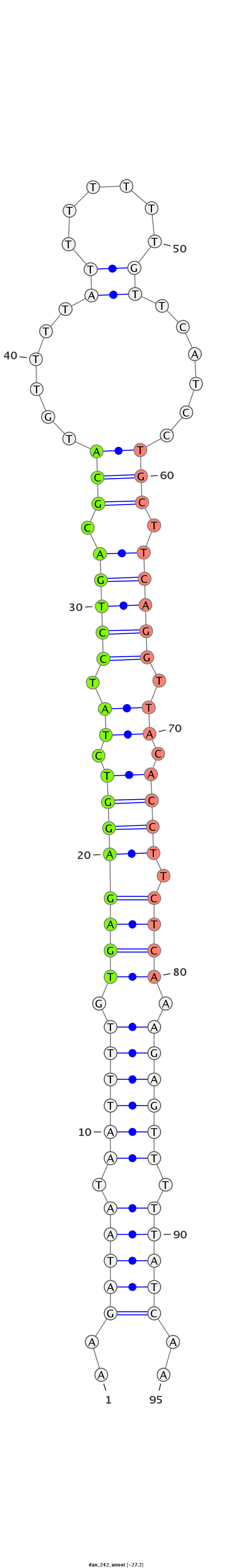
ID:dan_242 |
Coordinate:scaffold_12903:554769-554834 + |
Confidence:confident |
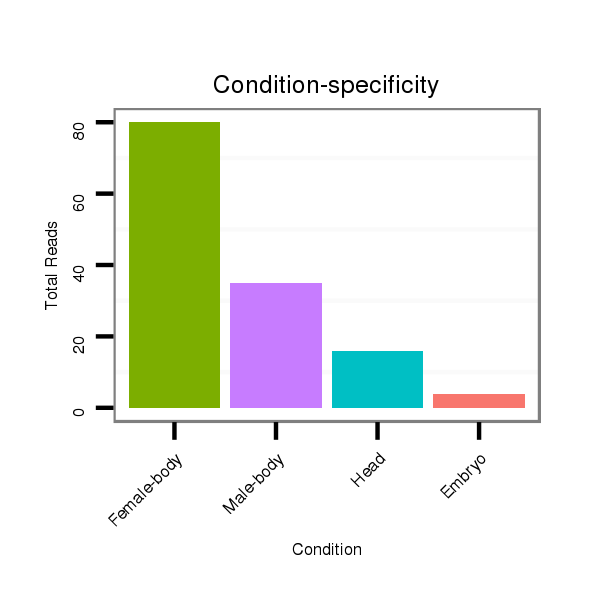
Class:Testes-restricted |
Genomic Locale:intergenic |
[View on UCSC Genome Browser {Cornell Mirror}] |
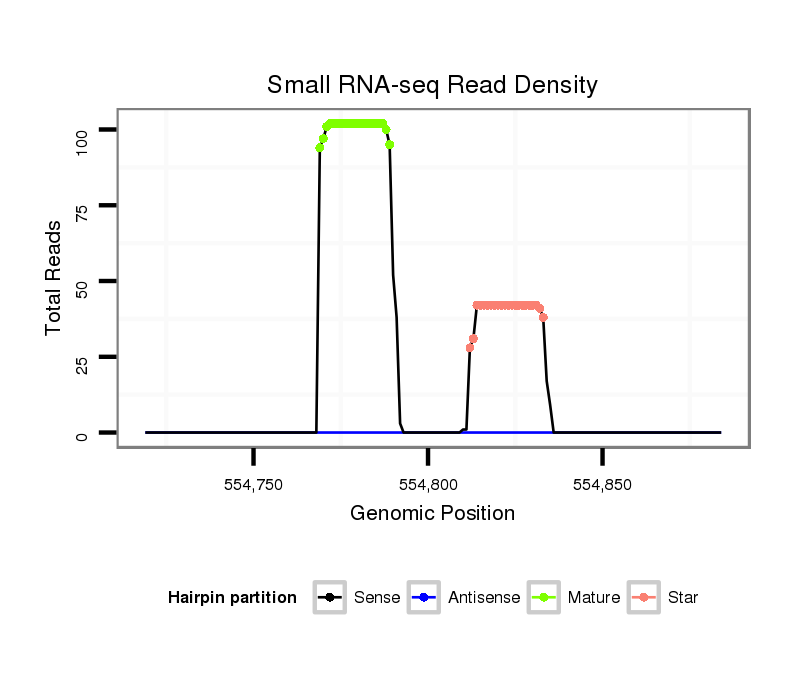
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -27.2 | -26.8 | -26.8 |
 |
 |
 |
intergenic
No Repeatable elements found
|
CAGTCATCATAGTGGTTTTTTTCAGTGTTTTTTTTAAGATAATAATTTTGTGAGAGGTCTATCCTGACGCATGTTTTATTTTTTTGTTCATCCTGCTTCAGGTTACACCTTCTCAAAGAGTTTTTATCAAAGCCTCGTTTATTTATATTTTAATTTTAAAATGAAA
***********************************..(((((.((((((.((((((((.((.(((((.(((......((......))......))).))))).)).))).))))).)))))).)))))..************************************ |
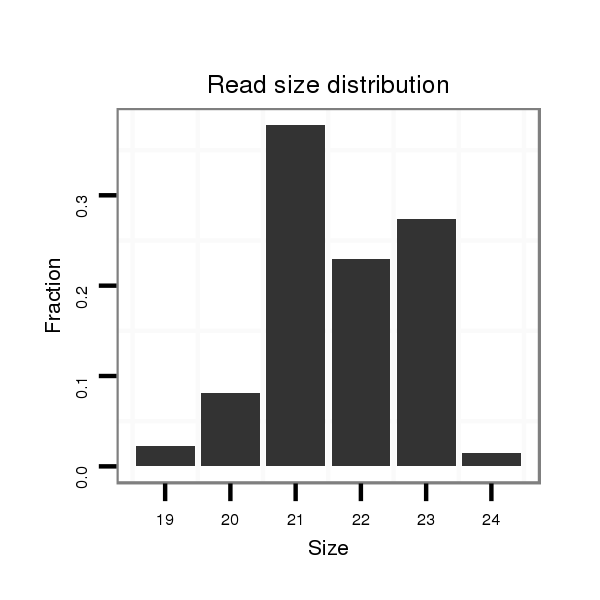
Read size | # Mismatch | Hit Count | Total Norm | Total | M044 female body |
V105 male body |
V106 head |
V055 head |
M058 embryo |
V039 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................TGAGAGGTCTATCCTGACGCA............................................................................................... | 21 | 0 | 1 | 40.00 | 40 | 18 | 15 | 4 | 2 | 1 | 0 |
| ..................................................TGAGAGGTCTATCCTGACGCATG............................................................................................. | 23 | 0 | 1 | 31.00 | 31 | 19 | 9 | 1 | 1 | 1 | 0 |
| .............................................................................................TGCTTCAGGTTACACCTTCTCA................................................... | 22 | 0 | 1 | 17.00 | 17 | 15 | 1 | 0 | 0 | 0 | 1 |
| ..................................................TGAGAGGTCTATCCTGACGCAT.............................................................................................. | 22 | 0 | 1 | 14.00 | 14 | 2 | 7 | 4 | 1 | 0 | 0 |
| ..................................................TGAGAGGTCTATCCTGACGC................................................................................................ | 20 | 0 | 1 | 5.00 | 5 | 2 | 0 | 1 | 2 | 0 | 0 |
| .............................................................................................TGCTTCAGGTTACACCTTCTCAA.................................................. | 23 | 0 | 1 | 5.00 | 5 | 5 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................CTTCAGGTTACACCTTCTCAAA................................................. | 22 | 0 | 1 | 5.00 | 5 | 3 | 0 | 2 | 0 | 0 | 0 |
| ....................................................AGAGGTCTATCCTGACGCATG............................................................................................. | 21 | 0 | 1 | 4.00 | 4 | 4 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................CTTCAGGTTACACCTTCTCA................................................... | 20 | 0 | 1 | 3.00 | 3 | 2 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................................TGCTTCAGGTTACACCTTCTCAAA................................................. | 24 | 0 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................CTTCAGGTTACACCTTCTCAA.................................................. | 21 | 0 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| ...................................................GAGAGGTCTATCCTGACGCA............................................................................................... | 20 | 0 | 1 | 3.00 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| ..................................................TGAGAGGTCTATCCTGACGCATGT............................................................................................ | 24 | 0 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................TGCTTCAGGTTACACCTTCTC.................................................... | 21 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..................................................TGAGAGGTCTATCCTGACG................................................................................................. | 19 | 0 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................TGAGAGGTCTATCCTGGCGCA............................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................GCTTCAGGTTACACCTTCTCT................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................TGAGAGGTCTATCCTGACGT................................................................................................ | 20 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .............................................................................................TGCTTCAGCTTACACCTTCTCA................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................TTAGAGGTCTATCCTGACGCA............................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..............................................................................................GCTTCAGGTTACACCTTCTCAAA................................................. | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................CCTGCTTCAGGTTACACCTTCTC.................................................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................TGAGAGGTATATCCTGACGCA............................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................TGAGAGCTCTATCCTAACGCA............................................................................................... | 21 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................TGAGAGGTCGATGCTGACGCCTG............................................................................................. | 23 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .............................................................................................TACTTCAGGTTACACCTTCTCA................................................... | 22 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................TGAGAGGTCTATCCTGACGCATGA............................................................................................ | 24 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................TGAGAGGTCTATCCTGACGCAA.............................................................................................. | 22 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................CGAGAGGTCTATCCTGACGCA............................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................TGGTTCAGGTTATACCTTCTCA................................................... | 22 | 2 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ..................................................TGAGAGGTCTATACTGACGCATG............................................................................................. | 23 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................TGAGAGGTGTATCCTGACGCATG............................................................................................. | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................TGAGAGGTCTATCCTGACGCATT............................................................................................. | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................TGAGGGGTCTATCCTGACGCATG............................................................................................. | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................GCTTCAGGTTACACCTTCT..................................................... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................TGAGAGGTATATCCTGACGCATG............................................................................................. | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................TCAGAGGTCTATCCTGACGCA............................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................GCTTCAGGTTACACCTTCTCA................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................GAGGTCTATCCTGACGCATGT............................................................................................ | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................TGAGAGATCTAGCTTGACGCA............................................................................................... | 21 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
|
GTCAGTAGTATCACCAAAAAAAGTCACAAAAAAAATTCTATTATTAAAACACTCTCCAGATAGGACTGCGTACAAAATAAAAAAACAAGTAGGACGAAGTCCAATGTGGAAGAGTTTCTCAAAAATAGTTTCGGAGCAAATAAATATAAAATTAAAATTTTACTTT
************************************..(((((.((((((.((((((((.((.(((((.(((......((......))......))).))))).)).))).))))).)))))).)))))..*********************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V039 embryo |
M058 embryo |
V105 male body |
|---|---|---|---|---|---|---|---|---|
| ..CAGTAGTTTCACCAAAAAAA................................................................................................................................................ | 20 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ...........CACCAAAAAAAGTCCCAACAAAA.................................................................................................................................... | 23 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 |
| ..CAGTAGTTTCAACAAAAAAA................................................................................................................................................ | 20 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 |
| ..........CCACCATAAAAAGTCACAAA........................................................................................................................................ | 20 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 |
| ............ACCAAAAAAACTCACAAAAA...................................................................................................................................... | 20 | 1 | 3 | 0.33 | 1 | 0 | 1 | 0 |
| ...........................AAAAAAAATTCTATCATTA........................................................................................................................ | 19 | 1 | 4 | 0.25 | 1 | 0 | 1 | 0 |
| ............ACCAAAAGAAGTCACAACAA...................................................................................................................................... | 20 | 2 | 5 | 0.20 | 1 | 0 | 1 | 0 |
| .........................................................................................TAGGAAGGAGTCCAAGGTGG......................................................... | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 1 |
| ..CAGTGGTTTCACCAAGAAAA................................................................................................................................................ | 20 | 3 | 9 | 0.11 | 1 | 1 | 0 | 0 |
| .............CCAAAGAAAGCCACAAAAAAAAC.................................................................................................................................. | 23 | 3 | 11 | 0.09 | 1 | 0 | 1 | 0 |
| ................AAAAAAGTCACCAAAAAA.................................................................................................................................... | 18 | 1 | 12 | 0.08 | 1 | 0 | 1 | 0 |
| ..............CAAAAAAAGTCACAAAAAC..................................................................................................................................... | 19 | 1 | 12 | 0.08 | 1 | 0 | 1 | 0 |
| .............CCACAAAAACTCACAAAAAAA.................................................................................................................................... | 21 | 2 | 14 | 0.07 | 1 | 0 | 1 | 0 |
| .............CCAAAAAAAATCACAAAAA...................................................................................................................................... | 19 | 1 | 16 | 0.06 | 1 | 0 | 1 | 0 |
| ........................................................................................CTAGGAAGGAGTCCAATG............................................................ | 18 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 |
| Species | Coordinate | ID | Type | Alignment |
|---|---|---|---|---|
| droAna3 | scaffold_12903:554719-554884 + | dan_242 | confident | CAGTCATCATAGTGGTTTTTTTCAGTGTTTTTTTTAAGATAATAATTTTGTGAGAGGTCTATCCTGACGCATGTTTTATTTTTTTGTTCATCCTGCTTCAGGTTACACCTTCTCAAAGAGTTTTTATCAAAGCCTCGTTTATTTATATTTTAATTTTAAAATGAAA |
| droBip1 | scf7180000395912:23739-23752 + | TAACCATCATAATG-------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
Generated: 10/20/2015 at 07:28 PM