
ID:dan_2377 |
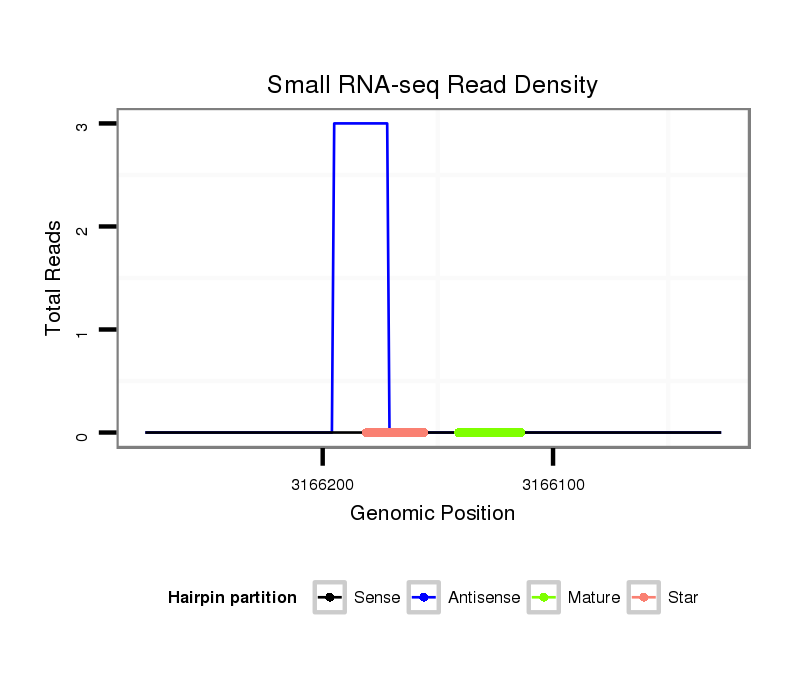
Coordinate:scaffold_13337:3166077-3166227 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -13.6 | -13.0 | -12.6 |
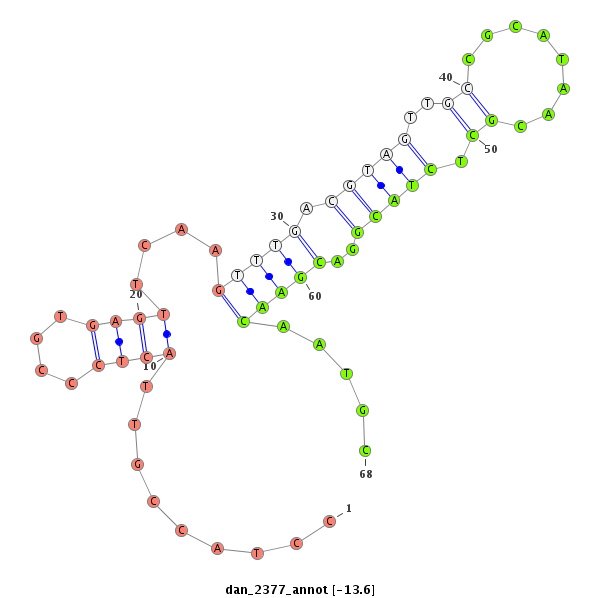 |
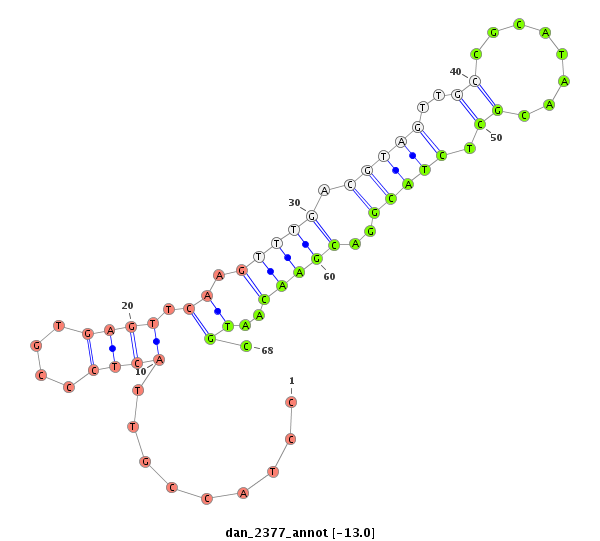 |
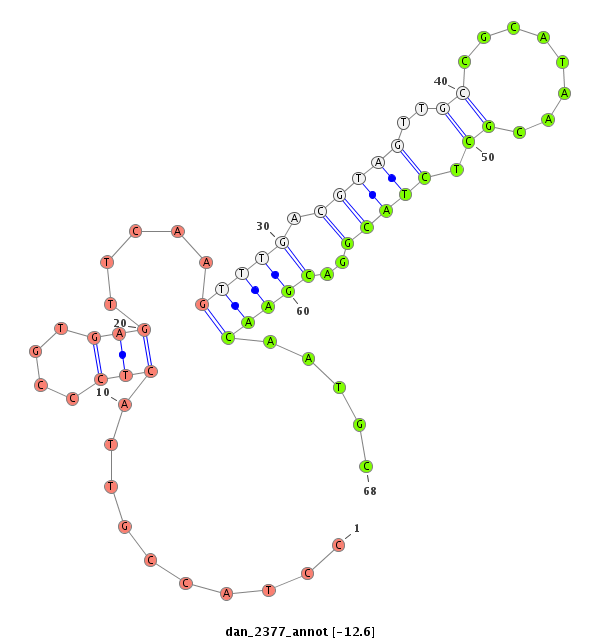 |
CDS [Dana\GF24704-cds]; exon [dana_GLEANR_9407:1]; intron [Dana\GF24704-in]
| Name | Class | Family | Strand |
| AT_rich | Low_complexity | Low_complexity | + |
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- TGATCACAGCAAGCGTCTGGCCAACCCGAGCAGTCCCTTGCAGAATCCAGGTAAATTTAGTCAGGACTATTTACGGTTTTGACTACGACTCCGGGACCTACCGTTACTCCCGTGAGTTCAAGTTTGACGTAGTTGCCGCATAACGCTCTACGGACGAACAATGCCATTTGAAAATATTCTTGTTACCTTTAATATTAAGAAATAATAGTAAATAAAATAATAACTTAAAAATTTTTATGGACGTTTATATT ************************************************************************************************......((.................(((((.(((((..((........)).)))))..)))))...))*************************************************************************************** |
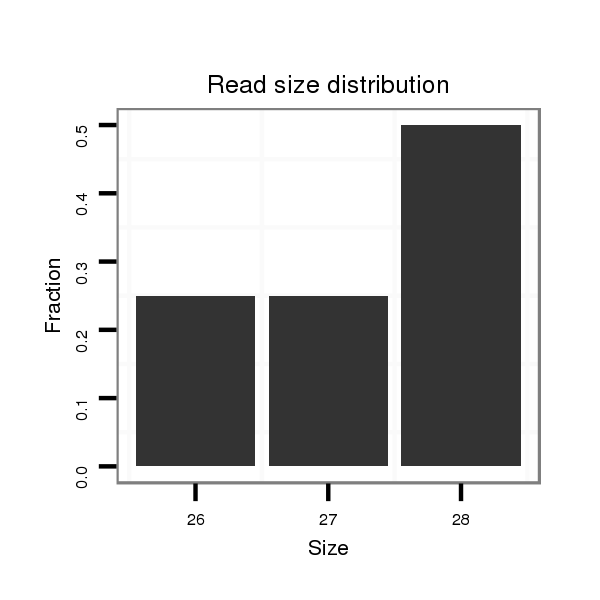
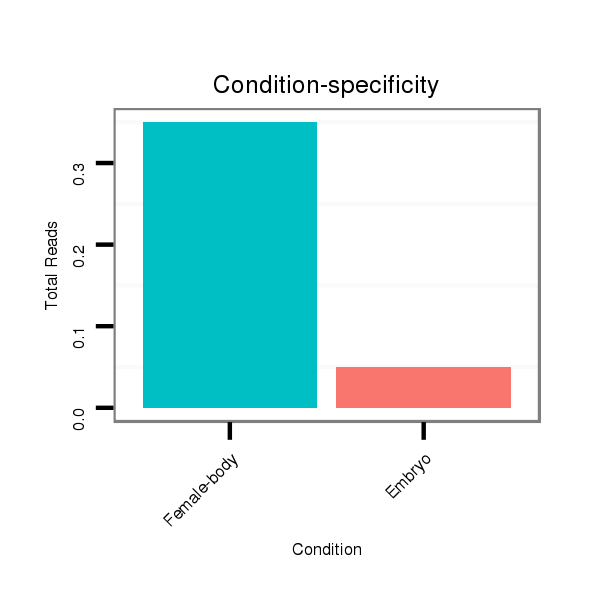
Read size | # Mismatch | Hit Count | Total Norm | Total | M044 female body |
M058 embryo |
V106 head |
|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................CGCATAACGCTCTACGGACGAACAATGC....................................................................................... | 28 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 |
| .GATCACAGCAAGCGTCTGGCCAACCC................................................................................................................................................................................................................................ | 26 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 |
| ...................................................................................................................................................CTACGTACCAAGAATGCCA..................................................................................... | 19 | 3 | 18 | 0.06 | 1 | 1 | 0 | 0 |
| .........CAAGCGTCTGGCCAACCCGAGCAGTCCC...................................................................................................................................................................................................................... | 28 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| ................................................................................................CCTACCGTTACTCCCGTGAGTTCAAG................................................................................................................................. | 26 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 |
| .........................................................................CGGTTTTGACTACGACTCCGGGACCTA....................................................................................................................................................... | 27 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| .....................................................................TTTACGGTTTTGACTACGACTCCGGGA........................................................................................................................................................... | 27 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| .....................................................................CGTACGGTTTAGACTACGA................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| .....................CAACCCGAGCAGTCCCTTGCA................................................................................................................................................................................................................. | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| .......................................................................................ACTCCGGGACCTACCGTTACTCCCGT.......................................................................................................................................... | 26 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
|
ACTAGTGTCGTTCGCAGACCGGTTGGGCTCGTCAGGGAACGTCTTAGGTCCATTTAAATCAGTCCTGATAAATGCCAAAACTGATGCTGAGGCCCTGGATGGCAATGAGGGCACTCAAGTTCAAACTGCATCAACGGCGTATTGCGAGATGCCTGCTTGTTACGGTAAACTTTTATAAGAACAATGGAAATTATAATTCTTTATTATCATTTATTTTATTATTGAATTTTTAAAAATACCTGCAAATATAA
***************************************************************************************......((.................(((((.(((((..((........)).)))))..)))))...))************************************************************************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | M044 female body |
V039 embryo |
M058 embryo |
V105 male body |
V106 head |
|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................GATGCTGAGGCCCTGGATGGCAAT................................................................................................................................................. | 24 | 0 | 20 | 3.40 | 68 | 68 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................GCTTGTTAAGGTAGACTTT.............................................................................. | 19 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................CAAGTTCAAACTGCATCAACGGCGT............................................................................................................... | 25 | 0 | 20 | 0.50 | 10 | 9 | 0 | 1 | 0 | 0 |
| ........................................................................................................................TCAAACTGCATCAACGGCGTATTGCG......................................................................................................... | 26 | 0 | 20 | 0.35 | 7 | 7 | 0 | 0 | 0 | 0 |
| .............................................................................................................................CTGCATCAACGGCGTATTGCGAGAT..................................................................................................... | 25 | 0 | 20 | 0.35 | 7 | 7 | 0 | 0 | 0 | 0 |
| ....................................................................................................GGCAATGAGGGCACTCAAGTTCAAACT............................................................................................................................ | 27 | 0 | 20 | 0.35 | 7 | 7 | 0 | 0 | 0 | 0 |
| ..................................................................................................................TCAAGTTCAAACTGCATCAACGGCGT............................................................................................................... | 26 | 0 | 20 | 0.35 | 7 | 7 | 0 | 0 | 0 | 0 |
| ................................................................................CTGATGCTGAGGCCCTGGATGGCAA.................................................................................................................................................. | 25 | 0 | 20 | 0.30 | 6 | 6 | 0 | 0 | 0 | 0 |
| .CTAGTGTCGTTCGCAGACCGGTT................................................................................................................................................................................................................................... | 23 | 0 | 20 | 0.20 | 4 | 0 | 0 | 4 | 0 | 0 |
| .........GTTCGCAGACCGGTTGGGCTCGTC.......................................................................................................................................................................................................................... | 24 | 0 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 | 0 |
| ...............................................................................TCTGATGCTGAGGCCCTGGATGGCAAT................................................................................................................................................. | 27 | 1 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 | 0 |
| ..........................................................................................................GAGGGCACTCAAGTTCAAACTGCATCA...................................................................................................................... | 27 | 0 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 | 0 |
| ..........................................................................................................GAGGGCACTCAAGTTCAAACTGC.......................................................................................................................... | 23 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 |
| ..........................GCTCGTCAGGGAACGTCTTAGGTCCA....................................................................................................................................................................................................... | 26 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 |
| .............................................................................................CCTGGATGGCAATGAGGGCACTC....................................................................................................................................... | 23 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 |
| ..........................................................................................................GAGGGCACTCAAGTTCAAACTGCAT........................................................................................................................ | 25 | 0 | 20 | 0.10 | 2 | 0 | 0 | 2 | 0 | 0 |
| .............................................................................................................GGCACTCAAGTTCAAACTGCATCA...................................................................................................................... | 24 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 |
| ...................................................................................ATGCTGAGGCCCTGGATGGCAATG................................................................................................................................................ | 24 | 0 | 20 | 0.10 | 2 | 0 | 0 | 2 | 0 | 0 |
| ............................................................AGTCCTGATAAATGCCAAAAC.......................................................................................................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| .CTAGTGTCGTTCGCAGACCG...................................................................................................................................................................................................................................... | 20 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| ..............................................................TCCTGATAAATGCCAAAAC.......................................................................................................................................................................... | 19 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ........CGGTCGCAGACCGGTTGGGCTCGTCA......................................................................................................................................................................................................................... | 26 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................GGGCTCGTCAGGGAACGT................................................................................................................................................................................................................. | 18 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................GGCACTCAAGTTCAAACTGCATCAACGG.................................................................................................................. | 28 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...TGTGTCGTTCGCAGACCGGTTGGGCT.............................................................................................................................................................................................................................. | 26 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| .................................................CCATTTAAATCAGTCCTGATAAAT.................................................................................................................................................................................. | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................AAGTTCAAACTGCATCAACGGCGTAT............................................................................................................. | 26 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................GTCAGGGAACGTCTTAGGTCCA....................................................................................................................................................................................................... | 22 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............................TCAGGGAACGTCTTAGGTCCATTTAAA................................................................................................................................................................................................. | 27 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................CCAAAACTGATGCTGAGGCCCTGGAT....................................................................................................................................................... | 26 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| .........GGTCGCAGACCGGTTGGGCTCGT........................................................................................................................................................................................................................... | 23 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................AGGGAACGTCTTAGGTCCATTTAAAT................................................................................................................................................................................................ | 26 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................................TGAGGCCCTGGATGGCAATGAGTG............................................................................................................................................ | 24 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ......................TTGGGCTCGTCAGGGAACGTCTT.............................................................................................................................................................................................................. | 23 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ....................GGTTGGGCTCGTCAGGGAACGTCTTAG............................................................................................................................................................................................................ | 27 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................TGCCAAAACTGATGCTGAGGCCCT........................................................................................................................................................... | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................CTGCATCAACGGCGTATTGCGAGATGC................................................................................................... | 27 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................................................CATCAACGGCGTATTGCGAGATG.................................................................................................... | 23 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| .......................................................................................................................................GGCGTATTGCGAGATGCCTGCTTGT........................................................................................... | 25 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................GGCGTATTGCGAGATGCCTGCTTG............................................................................................ | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ACTAGTGTCGTTCGCAGACCGGTTGGGC............................................................................................................................................................................................................................... | 28 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........CGTTCGCAGACCGGTTGGGC............................................................................................................................................................................................................................... | 20 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................AGTCCTGATAAATGCCAAAACTGATGCT................................................................................................................................................................... | 28 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................CCCTGGATGGCAATGAGGGCACTCAA..................................................................................................................................... | 26 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| ............................TCGTCAGGGAACGTCTTAGGTCCATT..................................................................................................................................................................................................... | 26 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| .................ACCGGTTGGGCTCGTCAGGGAACGTCT............................................................................................................................................................................................................... | 27 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................CGGTTGGGCTCGTCAGGGAACGT................................................................................................................................................................................................................. | 23 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................GCACTCAAGTTCAAACTGCAT........................................................................................................................ | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........GTTCGCAGACCGGTTGGGCTCGTCAG........................................................................................................................................................................................................................ | 26 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................TGCTGAGGCCCTGGATGGCAAT................................................................................................................................................. | 22 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...................................................................................................................CAAGTTCAAACTGCATCAACGGCG................................................................................................................ | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
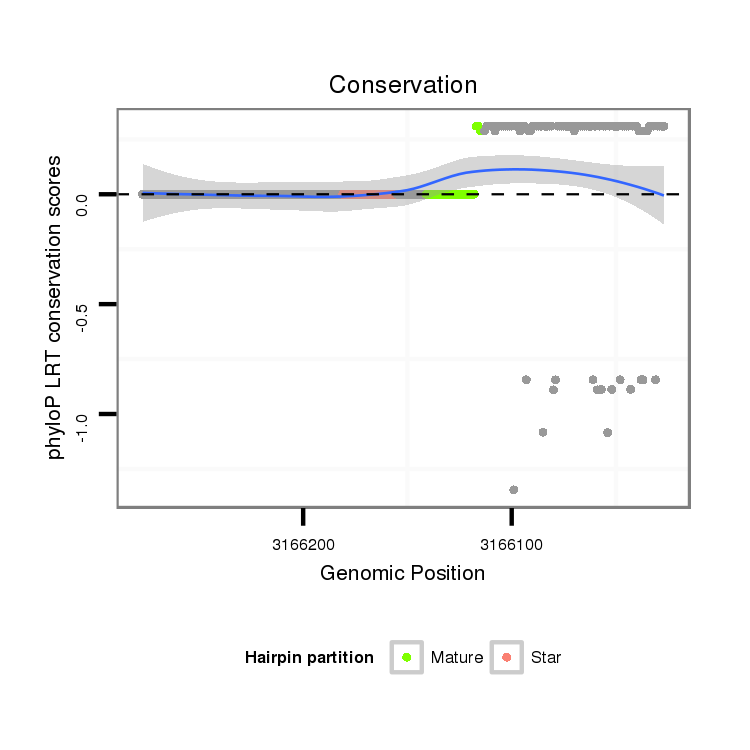
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droAna3 | scaffold_13337:3166027-3166277 - | dan_2377 | TGATCACAGCAAGCGTCTGGCCAACCCGAGCAGTCCCTTGCAGAATCCAGGTAAATTTAGTCAGGACTATTTACGGTTTTGACTACGACTCCGGGACCTACCGTTACTCCCGTGAGTTCAAGTTTGACGTAGTTGCCGCATAACGCTCTACGGACGAACAATGCCATTTGAAA----ATATTCTTGTTACCTTTAATATTAAGAAATAATAGTAAATAAAATAATAACTTAAAAAT------------------TTTTATGGACGTTTATATT |
| droBip1 | scf7180000395962:7120-7232 - | ----------------------------------------------------------------------------------------------------------------------------------------------------------------ATGCCATTTGAAAGTACATATTGTTGTTGCCTTTAAGATTATAAAATAATAGTAAATAAAGTTAAAAATAAAAGATATATACATAGAGCTTGGTTTATATGAGCGTTTGTATT |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
Generated: 05/18/2015 at 04:34 AM