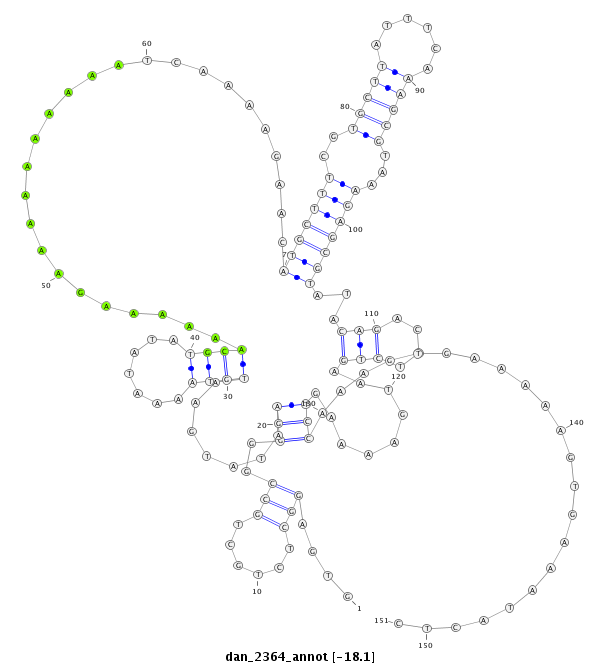




ID:dan_2364 |
Coordinate:scaffold_13337:2287715-2287865 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
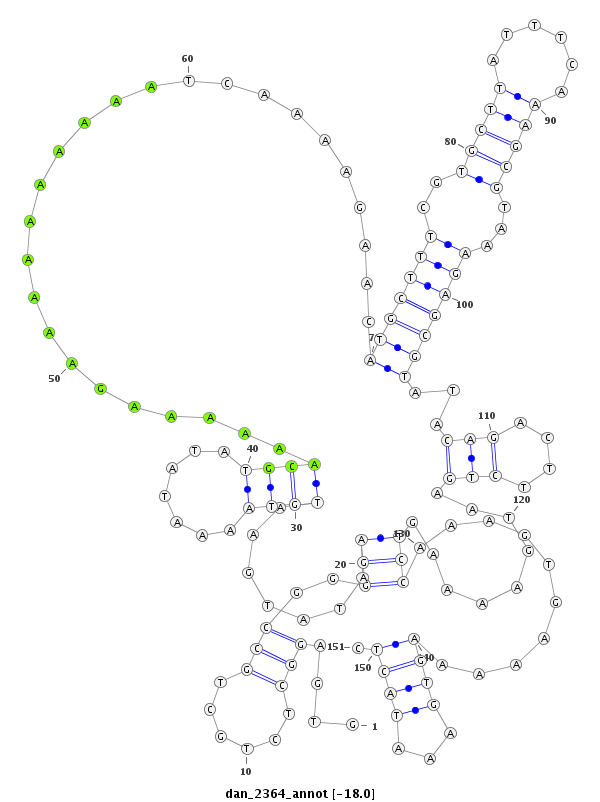
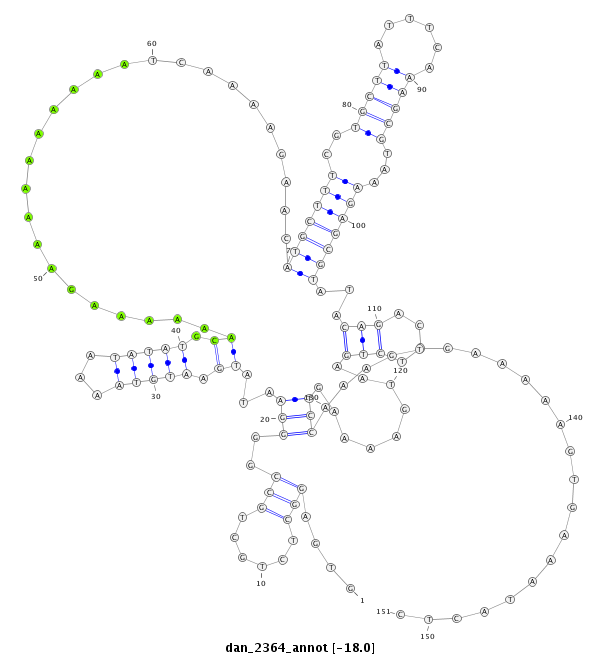
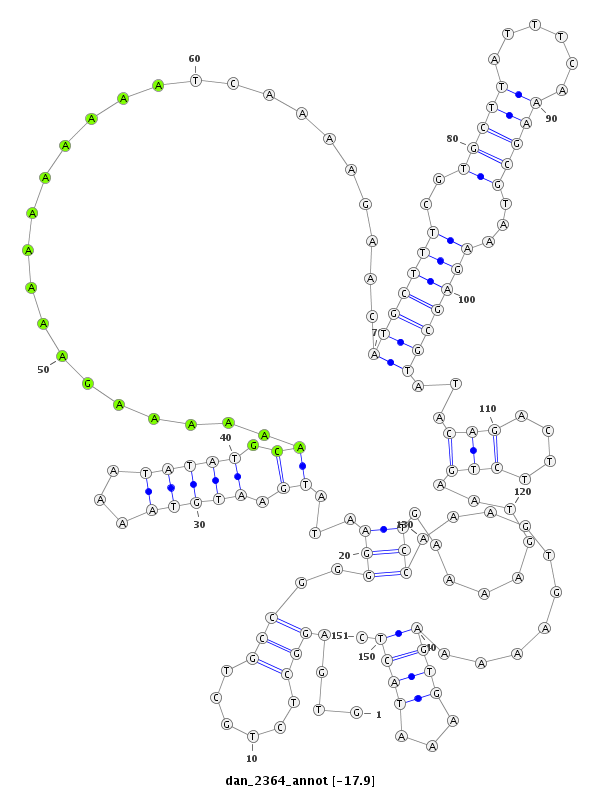
| -18.0 | -18.0 | -17.9 |
 |
 |
 |
exon [dana_GLEANR_9447:1]; CDS [Dana\GF24748-cds]; intron [Dana\GF24748-in]
No Repeatable elements found
| mature | star |
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- ATGCTGAGGCGCAGGCGCAGGGCGTGGCCCGGGAGGCTGAAGCTGAAGCTGTGAGGCTCTGCTGCCGGGGAATATGAATGTAAAATATATGCAAAAAAGAAAAAAAAAATCAAAAGAACATGCTTTCGTGCTTATTTCAAAGCGTAAAGAGCGTATACAGACTTCTGAATGAAAAGTCCAAAGTGAAAAAGTGAAATACTCGCACAACCTCCTGCGTGTAAGTTTTCAAGAGTTTTTTTTTTTTCAAAAGT **************************************************....(((......)))..(((.......((((.......))))..........................(((((((..(((((......)))))...)))))))...(((....))).........)))......................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M058 embryo |
V106 head |
V105 male body |
V055 head |
M044 female body |
V039 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................AAATATATGCTAAAAGGAAAAAA.................................................................................................................................................. | 23 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................GGAGGGTGAAGCTGATGC.......................................................................................................................................................................................................... | 18 | 2 | 13 | 1.00 | 13 | 0 | 12 | 1 | 0 | 0 | 0 |
| ...CTGAGGCGCATGCTGAGGGC.................................................................................................................................................................................................................................... | 20 | 3 | 6 | 0.83 | 5 | 0 | 0 | 4 | 0 | 1 | 0 |
| ...............................................................................................................................GTGCTTATTTGACAGCGT.......................................................................................................... | 18 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ............................CAGGGAGGGTGAAGCTGATGC.......................................................................................................................................................................................................... | 21 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................GGGAGGGTGAAGCTGATGC.......................................................................................................................................................................................................... | 19 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................AGGGAGGGTGAAGCTGATGC.......................................................................................................................................................................................................... | 20 | 3 | 10 | 0.30 | 3 | 1 | 2 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................GGAAAAAGAGAAAGACTCGCAC.............................................. | 22 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................GCAACAAAGGAAAAAAAAATCA........................................................................................................................................... | 22 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ............................CCGGGAGGCAGACGCTGA............................................................................................................................................................................................................. | 18 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...CTGAGGCGCATGCTCAGG...................................................................................................................................................................................................................................... | 18 | 2 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................GCAAAAAAGAAAAAAAAAA.............................................................................................................................................. | 19 | 0 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....TGAGGCGGAGGCGTAGGG..................................................................................................................................................................................................................................... | 18 | 2 | 16 | 0.13 | 2 | 0 | 0 | 0 | 0 | 2 | 0 |
| ............................CAGGGAGGGTGAAGCTGATG........................................................................................................................................................................................................... | 20 | 3 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................AAAAAAAAATCAAAAGACC.................................................................................................................................... | 19 | 1 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................AGAAAAAAAAAACGAAAAGAACA................................................................................................................................... | 23 | 2 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................AAAAAAAAACCAAAAGAACA................................................................................................................................... | 20 | 1 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................ATATGGGAAAAAGAAAAAAAAA............................................................................................................................................... | 22 | 2 | 19 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
|
TACGACTCCGCGTCCGCGTCCCGCACCGGGCCCTCCGACTTCGACTTCGACACTCCGAGACGACGGCCCCTTATACTTACATTTTATATACGTTTTTTCTTTTTTTTTTAGTTTTCTTGTACGAAAGCACGAATAAAGTTTCGCATTTCTCGCATATGTCTGAAGACTTACTTTTCAGGTTTCACTTTTTCACTTTATGAGCGTGTTGGAGGACGCACATTCAAAAGTTCTCAAAAAAAAAAAAGTTTTCA
**************************************************....(((......)))..(((.......((((.......))))..........................(((((((..(((((......)))))...)))))))...(((....))).........)))......................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V106 head |
M058 embryo |
M044 female body |
V039 embryo |
|---|---|---|---|---|---|---|---|---|---|
| ............................GTCCCTCCCACTTCGACTACG.......................................................................................................................................................................................................... | 21 | 3 | 3 | 12.00 | 36 | 23 | 5 | 8 | 0 |
| .............................TCCCTCCCACTTCGACTACG.......................................................................................................................................................................................................... | 20 | 3 | 10 | 7.90 | 79 | 76 | 1 | 2 | 0 |
| ............................GTCCCTCCCACTTCGACTAC........................................................................................................................................................................................................... | 20 | 3 | 10 | 2.40 | 24 | 8 | 5 | 8 | 3 |
| ..............................CCCTCCCACTTCGACTTC........................................................................................................................................................................................................... | 18 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| .................................................................................................................................................................................TGTTTCACTTTTTCACTTTA...................................................... | 20 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ..............................CCCTCCCACTTCGACTACG.......................................................................................................................................................................................................... | 19 | 2 | 3 | 1.00 | 3 | 3 | 0 | 0 | 0 |
| ..............................CCCTCCCACTTCGACTACGC......................................................................................................................................................................................................... | 20 | 3 | 18 | 0.44 | 8 | 5 | 2 | 1 | 0 |
| ............................................CTTGGACACTCAAAGACGACG.......................................................................................................................................................................................... | 21 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 |
| ............................GTCCCTCCCACTTCGACT............................................................................................................................................................................................................. | 18 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 |
| ............................GTCCCTCCCACTTCGACTCCG.......................................................................................................................................................................................................... | 21 | 3 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 |
| ..............................................................................................................................................................................................................................AAAAGATCTCAAAAAAAAAAA........ | 21 | 1 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 |
| .............................TCCCTCCCACTTCGACTGCG.......................................................................................................................................................................................................... | 20 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 |
| ..............................CCCTCCCACTTCGACTACGCC........................................................................................................................................................................................................ | 21 | 3 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 |
| .............................................................................................................AGTGATCTTGGACGAAAGC........................................................................................................................... | 19 | 3 | 16 | 0.13 | 2 | 2 | 0 | 0 | 0 |
| ...........................CGTCCCTCCCACTTCGACT............................................................................................................................................................................................................. | 19 | 3 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droAna3 | scaffold_13337:2287665-2287915 - | dan_2364 | ATGCTGAGGCGCAGGCGCAGGGCGTGGCCCGGGAGGCTGAAGCTGAAGCTGTGAGGCTCTGCTGCCGGGGAATATGAATGTAAAATATATGCAAAAAAGAAAAAAAAAATCAAAAGAACATGCTTTCGTGCTTATTTCAAAGCGTAAAGAGC---------------GTATACAGACTTCTGAATGAAAAGTCCAAAGTGAAAAAGTGAAATACTCGCACAACCTCCTGCGTGTAAGTTTTCAAGAGTTTTTTTTTTTTCAAAAGT |
| droBip1 | scf7180000396390:134220-134380 - | CTGCTGAGGCGCAGGCGCAGGGCGTGGC------TGCTG-------------------------GCGG-GAGCATGAATGCGAGATGTATGCAAAAC--------------------TCATGCCTTCGT-----------AGCGTACTCGGCGGATACACACGGCCCATACACACAGTTCTGAATGAA--CCCGCAAGTGTAC--------------------------GGCGTGAGTTCGGAGGA--------------AGAGGG | |
| droKik1 | scf7180000302510:879347-879396 + | GCTCGGAAGCGCAGGCGCAGGGCGTGGC------TGCTG-------------------------GGAA-AA-----------------------------------------------------------------------TAAA---------------------------------------------------------------------------------------------------G--------------GAAAAG | |
| droTak1 | scf7180000414364:19617-19640 + | -----GAAGCGCAGGCGCAGGGCGTGGCC--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| dp5 | XR_group8:1406757-1406807 - | -----GAGGCGCAGGCGCAGGGCGTGGTCCTGTCTGCTT-------------------------AGGA-AA-----------------------------------------------------------------------TTTA---------------------------------------------------------------------------------------------------A--------------AGTGTG | |
| droPer2 | scaffold_32:69380-69430 + | -----GAGGCGCAGGCGCAGGGCGTGGTCCTGTCTGCTT-------------------------AGGA-AA-----------------------------------------------------------------------TTTA---------------------------------------------------------------------------------------------------A--------------AGTGTG |
| Species | Read alignment | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||
| droPer2 |
|
Generated: 05/18/2015 at 04:27 AM