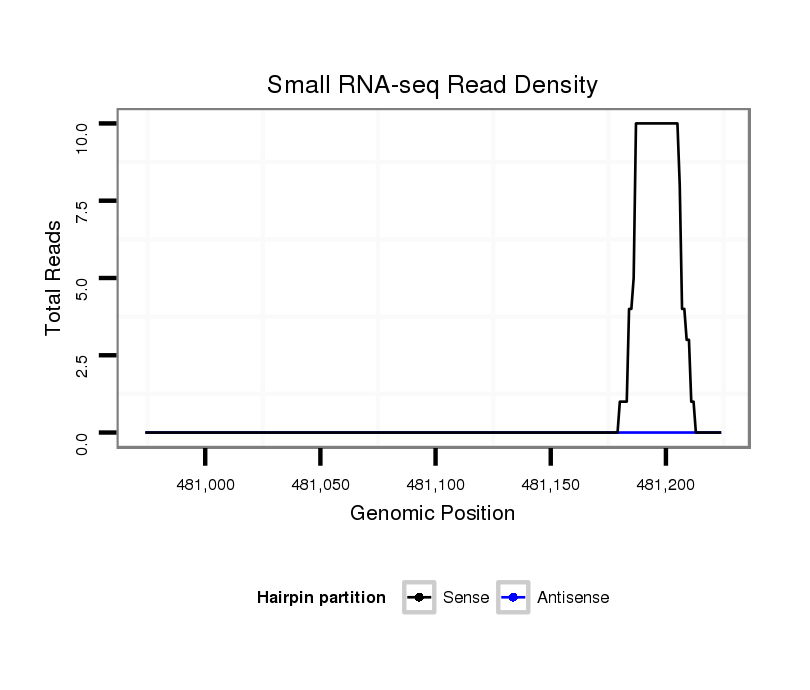
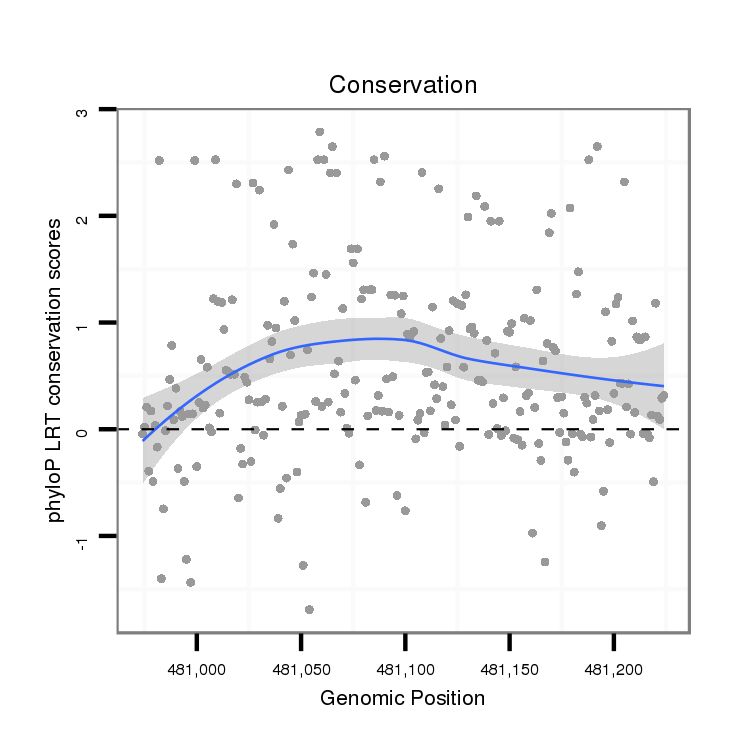
ID:dan_2317 |
Coordinate:scaffold_13337:481024-481174 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
exon [dana_GLEANR_9603:4]; CDS [Dana\GF24919-cds]; intron [Dana\GF24919-in]
| Name | Class | Family | Strand |
| (CAA)n | Simple_repeat | Simple_repeat | + |
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## TTGCAGCTGGGTTTCCCCAAAGCGCCCATATTTGCATTATGCAATCGAACCCCTTCACGCACACACCAACAACAGCAATAGCAACAGCAACAGCGACACCAGCAACACAAACCACAACAAGCCGCATTCATGTGCTTCAAAGTCTTACTATATCAACACGCATTTTCCACTCTCAAAAAACACAAACACCCAACTCATCAGCCAGCAGCAGCAGCAGCAGTCGAGGAAAAGCAGACATCGTATCTGCCGCT **************************************************.............................(((.(((......(((...........................))).....)))))).................................................................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M058 embryo |
V106 head |
V105 male body |
V039 embryo |
M044 female body |
V055 head |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................TTACTAGATCAACACTCATT....................................................................................... | 20 | 2 | 2 | 6.50 | 13 | 13 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................TTACTATATCCACACTCATT....................................................................................... | 20 | 2 | 3 | 2.67 | 8 | 8 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................GCAGCAGTCGAGGAAAAGCA.................. | 20 | 0 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................TTACTAGATCCACACTCATTTT..................................................................................... | 22 | 3 | 4 | 2.00 | 8 | 8 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................GCAGCAGCAGTCGAGGAAAAGCAGACA.............. | 27 | 0 | 1 | 2.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................CGCAAAAAACACAAACACC............................................................. | 19 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................GCAGCAGTCGAGGAAAAGCAGACATC............ | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................AGCAGCAGTCGAGGAAAAGCA.................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ....................................................................................................................................................................................................................CGCAGCAGTCGAGGAAAAGCA.................. | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................TTACTATATCCACACTCATTT...................................................................................... | 21 | 2 | 2 | 1.00 | 2 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................................................................................................................................AGCAGCAGCAGCAGTCGAGGAAAAGCA.................. | 27 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................GCAGCAGTCGAGGAAAAGC................... | 19 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................ATGTGCTTCATACTATTACTA..................................................................................................... | 21 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..................................................................................................................................................................................................................GCAGCAGCAGTCGAGGAAAAGC................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................GCAGCAGTCGAGGAAAAGCAGA................ | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................TTACTAGATCCACACGCATT....................................................................................... | 20 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................CCTCAGCCAGCAGCAGCAGCAGCAGT.............................. | 26 | 1 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................AGCAGCAGCAGACGAGGAA....................... | 19 | 1 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ................................................................................................................................................................................AAAACCCAAACACCCAACCCA...................................................... | 21 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................AAAAACACAAACACCCGACAC....................................................... | 21 | 2 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................ATCTAAAAAAACACAAACACCCC........................................................... | 23 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................GCCGTAGTCGATGAAAAGCAG................. | 21 | 3 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................TGACTAGATCAACACTCATTTT..................................................................................... | 22 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................AAAAAAACACAAACACCCAA.......................................................... | 20 | 1 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................CAAAAAACACAACAACCCAAC......................................................... | 21 | 2 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................TTACTATATCCACACTCAT........................................................................................ | 19 | 2 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................TGACTATATCAACACTCGTT....................................................................................... | 20 | 3 | 6 | 0.17 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ................................................................................................................................................................................AAAACACAAACACCCAAACCA...................................................... | 21 | 2 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................TGTGCTTCATACTCATACTA..................................................................................................... | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ................................................................................................................................................TTGCTATATCCACACTCATTT...................................................................................... | 21 | 3 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................GCAACAGAGACACGAGCCACA................................................................................................................................................ | 21 | 3 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................TTACTAGATCAACACTCAT........................................................................................ | 19 | 2 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................AAAAAACCCAAACTCCCAAC......................................................... | 20 | 2 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........................................................................................................ACACAAACCATAAAAAGCC................................................................................................................................ | 19 | 2 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................TACCAACTCATCAGCCAG.............................................. | 18 | 2 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...........................................................................................................................................AACTCTTACTCGATCAACAC............................................................................................ | 20 | 3 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................AAACAACAGCAATAGCAACAGCAAATG.............................................................................................................................................................. | 27 | 3 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................................................GAAACACAAACACCCAACAC....................................................... | 20 | 2 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................TTACTATATCCACACCCCTT....................................................................................... | 20 | 3 | 11 | 0.09 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .....................................................................................................................................................................................ACAAACACCCAACCCACCA................................................... | 19 | 2 | 12 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .................................................................................................................................................................................AAACACAAACACCCAAACCA...................................................... | 20 | 2 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................AAAAACAAACACCCAACCCA...................................................... | 20 | 2 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................TTACTAGATCAACACTCATC....................................................................................... | 20 | 3 | 14 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................TTACTATATCCACACTCTTT....................................................................................... | 20 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .................................................................................................................................................................................AAACACGAACACCCAACC........................................................ | 18 | 2 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................CAAAATACCCAAACACCCA........................................................... | 19 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................AACAAAAACACCCAACCC....................................................... | 18 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................AGCAGCAGCAGCAGCA................................ | 16 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| .............................................................................................................................................................................CAAAAAAAACTAACACCCCAC......................................................... | 21 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................CAAAAAAAACAAAAACGCAACTC....................................................... | 23 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................TTAACGCAGACACCAAGAA................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................CAGCAGTCGCGGTAAAACA.................. | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................ATCAAAAAAAACAAACACCC............................................................ | 20 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
|
AACGTCGACCCAAAGGGGTTTCGCGGGTATAAACGTAATACGTTAGCTTGGGGAAGTGCGTGTGTGGTTGTTGTCGTTATCGTTGTCGTTGTCGCTGTGGTCGTTGTGTTTGGTGTTGTTCGGCGTAAGTACACGAAGTTTCAGAATGATATAGTTGTGCGTAAAAGGTGAGAGTTTTTTGTGTTTGTGGGTTGAGTAGTCGGTCGTCGTCGTCGTCGTCAGCTCCTTTTCGTCTGTAGCATAGACGGCGA
**************************************************.............................(((.(((......(((...........................))).....)))))).................................................................************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V055 head |
M044 female body |
V105 male body |
M058 embryo |
V106 head |
|---|---|---|---|---|---|---|---|---|---|---|
| ...............................................................................................TGTGGTCGATTGGTTTGGTGTT...................................................................................................................................... | 22 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..CGTCGACCCAAAGGGGTAAG..................................................................................................................................................................................................................................... | 20 | 3 | 12 | 0.25 | 3 | 1 | 2 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................TTCGTCTATGGGATAGACGGC.. | 21 | 3 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................TTTTGCTGGTATGAACGTAA..................................................................................................................................................................................................................... | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 1 | 0 | 0 |
| .................................................TAGGAAGTGCGTGTGTTGTT...................................................................................................................................................................................... | 20 | 3 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................TCGTCCGTAGCAAAGACGGA.. | 20 | 3 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................GGGTATAGATGTAATACGATAG............................................................................................................................................................................................................. | 22 | 3 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................GTGTTTGCGGCTTGAGGAGT................................................... | 20 | 3 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................GCGGTGGTCGTTGTGTGGGGT......................................................................................................................................... | 21 | 3 | 16 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 |
| ....................................AATACGTTAGCTAGAGGCA.................................................................................................................................................................................................... | 19 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droAna3 | scaffold_13337:480974-481224 + | dan_2317 | TTGCAGC---TGG--------------------G--T-TT-----C-CCC----AAAGC--------GC-------CC--ATATTTG------CATT-ATGCAATCGAAC--CCCTTCACGCA-----CACA---------------CCAA-----C--------------AACAGCAATAGCAACAGCAA---CAGCGACAC------CAGCAACACAAACCACAA--CAAGCCGC------ATT--CATGTGCTTCAAAGTCTTACTA---TATCAA-------------------------CACGCATTTTCCACTCTCAA-A--A-AACACAA------------------------------------------------------------------ACA--C----------C-CAACTCATCAGCCAGCA--G----------------------CAG----CAGCAGCAGT---CGAGGAA-------AAGCAG--ACAT------CGTATCTGCCGCT |
| droBip1 | scf7180000395155:278189-278446 - | TTGCACC---TGG--------------------G--T-TTCC---C-CTT----AAGCT--------TC-------CC--ATATTTG------CATT-ATGCAATCTCGAACCCCTTCACGCT-----CACA---------------CAC------C------ACC--AACATCAGCTACAGCAACACCAG---CAACAGAAA------------------CCACAA--CAAGCTGC------ATT--CATATGCTTTAAAGTCTTACTA---TATCAA-------------------------CACGCATTTTCCA-TCCTCA-ACCA-AACACAA------------------------------------------------------------------ACA--C----------C-CAACTCATCAGCCACCA--CCA----------------GCAGCAT----TAGCAGCAGTCTCCGAAGAA-------AAGCAG--ACAT------CGTATCTGCCGCT | |
| droKik1 | scf7180000302441:1307496-1307714 - | TTTGAGATCTTGAGT---CTTAAGTT---TTGAGTCT-TTTTCTTC-C----------------------------CC------TTG--TGATTATT-ATGCAATCGAAA--CCTTTCACGAA-A----------------------------------------------A--------ACAAACAGCA-----------AA------------------CCACAACACAAGCCGC------AAT--CATATGCTTTAATGTCTTT--ACTATATCAACAAAACAAAATCAAACAAAAAACTCCACGCATT---------------------ACAA-----------------------------------------------------------------------AAA--------------------------A--ATA---------ACAGCCAG---CAG----CAGCCAAAGA---GGAGGAA-------AAGCTG--GCA-------------------- | |
| droFic1 | scf7180000454105:769289-769560 + | TTGGAT----TGAGTTGGCCTGAGCTGAGCTGAG--T-TTTT---C-TCC----CAGAC--------CC-------CCTTATATTTG------CATT-ATGCAATCGACAA-CCGTTCACGCA-----CACA---------------AAAA-----G-----------------AAACGACACAGCAGCAG---CAGCAGCAA------------------CCACAA--CAAGCGGC------AAT--CATAAGCTTTAATGTCTTACTA---AAACAA------------------CAAACTCCACGCATTTTACCCTCTTGA-A--ATCACACAA----------------------------------------------------------------AACAA--AAAA-ACCAAAC-CCAAACATCAAAATGCA--ACA----------------A---CAGCCAACAGCCAAAGC---CGAGGAA-------AAGCTG--GCA-------------------- | |
| droEle1 | scf7180000491249:2976395-2976659 - | GTGAAGT---TGAGT-----TAA-----GTTGAG--T-TTTT---C-TCG----CAGAC--------AC-------CC--ATATTTG------CATT-ATGCAATCTCAAA-CCATTCACGCA-----CACA---------------CAA------A-----------------AATAACAACAACAGCAAAAGCAGCAGCAA------------------CCACAA--CAAGCAGC------AAT--TATAAGCTTTAATGTCTTACTA---AAACAA------------------CAAACTTCACGCATTTTACCCTCTTAA-A--A-AATATAA-------------------------------------------------------------------------AAAAAAAAACACAAAACATCAAAATGCAACACAACAGCCACA-------G---CGAA---CAGCCAAAGT---CGAGGAA-------AAGCTG--GCA-------------------- | |
| droRho1 | scf7180000776529:347857-348072 + | TCGCGAT---TGGA-------------------G--C-C----------------AATG--------CG-------CG--GTATTTCCATA---ATC-ATTGAATCGCCAG-AAGTG-----------------------------------GCCAC---AGCC--AC---A------GCAGCAACAGCAA---CAGCAACGGCAACACAAGTAGCAGCAACTGCCA--AGAGCAAC------AG------CAACACTGAAATGTCGCTC---AAGTCC------------------CAGGCT---GGCATGTTGCTGGC--------A-TTTGG------------------------------------------------------------------------------------------------CGACACA--GCA----------------G---CAA----CAACCAAAGA---TAAT-----TTGG-AAACTA--GTT------------------AC | |
| droBia1 | scf7180000302428:9207469-9207704 + | TTGGAGC---AGA--------------------G--T-TTTT---C-TCC--CATAGAC--------CC-------CC--ATATTTG------CATT-ATGCAATCCAAAAACCATTCACGCA-----CACA---------------CCAA-----A-----------AACAACAGCAACAGCAACATCAA---CAGCAGCAA------------------CCACAA-ACAAGCAGC------AAT--CATAAGCTTTAATGTCTTACTA---AAACAA------------------CAAACTCCACGCACTTTACCCTCTTAA-A--A-AATACAA---------------------------------------------------------------CAAAAT--A----------C-G---------------AAAACA----------------A---CAGC---CAGCCAAAGT---CGAGGAA-------AAGCTG--GCA-------------------- | |
| droTak1 | scf7180000415857:189755-189993 + | TTGGAGC---TGC--------------------G--T-TTTT---C-TCC----CAGGCCCCCCCCCTC-------CT--ATATTTG------CATT-ATGCAATCCCAAA-CCATTCACGCA-----CACA---------------CCAA-----A--------------AACAACAACAGCCACAGCAG---CAGCAGCAA------------------CCACAA--CAAGCAGC------AAT--CATAAGCTTTAATGTCTTACTA---AAACAA------------------CAAAATCCACGCATTTTACCCTCTTAA-A--A-AATACAAAAA-----------------------------------------------------------------------------------------CAAAATACAAAACA----------------A---CAGC---CAGCCAAAGT---CGAGGAA-------AAGCTG--GCA-------------------- | |
| droEug1 | scf7180000409466:172255-172514 + | TTGGAGC---TGA--------------------C--T-TTTCCT---TCC----CAGAC--------CC-------CC--ATATTTG------CATT-ATGCAATCCCAAACCCATTCACGCA-----CACA---------------CCAAAACCAACACAGCA--CT---AACAGCAGCAGCAACAGCAA---CAGCAGCGA------------------CCACAA--CAAGCAGC------AAT--CATAAGCTTTAATGTCTTACTA---AAACAA------------------CAAACTCCACGCATTTTACCCTCTTAA-A--A-AAC----------------------------------------------------------------CAACAACAC--AAAA-------C-CAAAACATCAAAATGCA--ACA----------------G---CAGC---CAGCCAAAGT---CGTGGAA-------AAGCTG--GCA-------------------- | |
| dm3 | chr3L:2442244-2442465 + | TTGGAGC---CGA--------------------GTTTTTTTT---C-CCC----CAGAC--------AC-------CC--ATATTTG------CATT-ATGCAATCCAAAA-CC---------AA----------------------------------------------A------ACAACAACAGCAA---CAGCAGCAA------------------CCACAA--CAAGCAGC------AAT--CATAAGCTTTAATGTCTCACTA---AAAC-------------------------TCCACGCATTTTACCCTCCTAA-A--A-AATAA-----------------------------------------------------------TAACAACAACAAAAAAAA-----------ACACATCAAAATGCT--ACA----------------G---CAGA---GAGCCAAAGT---CGAGGAA-------AAGCTG--GCA-------------------- | |
| droSim2 | 3l:2339009-2339219 + | TTGGAGC---CGA--------------------G--T-TTTT---C-CCC----CAGAC--------AC-------CC--ATATTTG------CATT-ATGCAATCCAAAA-CC---------AA-------------------------------------------------------AACAACAGCAA---CAGCAGCAA------------------CCACAA--CAAGCAGC------AAT--CATAAGCTTTAATGTCTCACTA---AAAC-------------------------TCCACGCATTTTACCCTCCTAA-A--A-AATGA--------------------------------------------------------------CAACAACAA--AAAA-----------ACACATCAAAATGCT--ACA----------------G---CAGA---CAGCCAAAGT---CGAGGAA-------AAGCTG--GCA-------------------- | |
| droSec2 | scaffold_2:2473195-2473402 + | TTGGAGC---CGA--------------------G--T-TTTT---C-CCC----CAGAC--------AC-------CC--ATATTTG------CATT-ATGCAATCCAAAA-CC---------AA-------------------------------------------------------AACAACAGCAA---CAGCAGCAA------------------CCACAA--CAAGCAGC------AAT--CATAAGCTTTAATGTCTCACTA---AAAC-------------------------TCCACGCATTTTACCCTCCTAA-A--A-AATAA-----------------------------------------------------------------CAACAA--AAAA-----------ACACATCAAAATGCT--ACA----------------G---CAGA---CAGCCAAAGT---CGAGGAA-------AAGCTG--GCA-------------------- | |
| droYak3 | 3L:9492182-9492418 - | dya_752 | TTGGAGC---TGC--------------------T--T-TTTT-T-C-CCCCCCAAAGAC--------AC-------CC--ATATTTG------CATT-ATGCAATCCAAAA-CC---------AA------A---------------ACAAAACCAA--------------AACAGCACCAACAACAGCAA---CAGCAGCAA------------------CCACAA--CAAGCAGC------AAT--CATAAGCTTTAATGTCTCACTA---AAACAAC---------------AACAAACTCCACGCATTTTACCCTCCTAA-A--A-AATACAA-------------------------------------------------------------------------AA-----------ATACATCGAAATGCA--ACA----------------G---CAGC---CAGCCAAAGT---CGAGGAA-------AAGCTG--GCA-------------------- |
| droEre2 | scaffold_4784:2458208-2458431 + | der_699 | TTGGAGC---TGA--------------------G--T-TTTT---C-CCC----CAGAC--------AC-------CC--ATATTTG------CATT-ATGCAATCCAAAA-CC---------AA--------------------------AACCAA--------------AACAACAGCAACAACAGCAA---CAGCAGCAA------------------CCACAA--CAAGCAGC------AAT--CATAAGCTTTAATGTCTCACTA---AAACAA------------------CAAACTCCACGCATTTTACCCTCCTAAAA--A-AATACAA-------------------------------------------------------------------------AA-----------ATACATCAAAATGCT---CA----------------G---CAGC---CAGCCAAAGT---CGAGGAA-------AAGCTG--GCA-------------------- |
| dp5 | XL_group1e:9035951-9036163 + | GCAG--C---TGC--------------------A--------------------ATAAT--------GC------ACA--TTTCTGCTGAGGTCA---GTTCGACC------ACCTCAAATCAAA---TACAGC-----------------------------A--GC---AACAGCAACAGCAACAACAA---CAGCAGCAG------CAGCAG------CACCAA--CA-GCTTCACCAACAG------TTGCA-------CCAACAA---CACCAA-------------------------------------------------------------------------------------------------------------------------CAGCAA--ACGA-------C-TCCAACAGCAACG-ACC--ACA---------ACGGGCGG---CAG----CACCACGAAG---CGGCGTAATGCAG-AGTC--------------------CTCCTCC | |
| droPer2 | scaffold_33:432649-432848 + | --------------------------------------------------------------------------------------G------CATT-ATGCAATCGAAAA-----------------TAC----------------------------------------AACAGCAACAACAACAACAA---CAACAACAA------------------CAACAA--CAACAAGA------AATCCCACATGCTTTAATGTCT--------------------------------------CCACGCATT-TACCCCCTCAA-A--A-AAAACACATGCAACATGCAACATGCAGCA-------------------------------------GCAGCAGCAGCAACA------------------------ACA--GCA------ACAACAAGCAG---CAG----CCGCAACAGC---AGGGGAA-------AAGCTC--CCA------------------CC | |
| droWil2 | scf2_1100000004515:2943635-2943846 + | CAGAAAG---AGG--------------------A--A-TT-----C-CTC----AAATC--------GCCAGTACCCA--GTAACGG----ATCACC-AGGCAATCAACAG-CA---------AC----------------------------------------------A---GCAGCAACAGCAGCAA---CAGCAGCAA---CACCAGCAACAACAACAA---------CA------------------ACATTTCTTTCCTACAA---CTTCAA-------------------------------------------------C-AGTACACAATCAATTTCCAGCA--CAACCGGCGCTACCGCCACGAGGA----------------CGCCAACAGCA------------------------------GCA--ACT------GCA-------G---CAAG---CAGC---AAT------------------------------------------------ | |
| droVir3 | scaffold_13246:1792488-1792701 - | TATCGC-----GAGT------------------G--T-TAGT---G-CCA----ATAAC--AACAACAA-------CA--ATGTGAA------C---------------------TTAACGAA-----TGCAACTGCAACAGCAATAACA--ACAAC--------------AACAGCAACAACAGCAGCTA---CAGCTGCAA------------------CAGCAA--C-AGCAGC------GG------AAGCA-------------------ACGA------------------CAAC-ATCGCTCGGCT--------TGA-G--C-GATGAAC----------------------------------------------------------------AGCAA--CAAT-----------CGACATCGGC----A--GCA----------------G---CAG----CAGCAGCAGC---AGCAGCA-------A--CTG-----------------TTGCTGCC | |
| droMoj3 | scaffold_6473:5472848-5473118 + | CTGCAGC---AGC--------------------T--C-C--------GCA----AATGC--------AT-------CC--ACATGCG------CATGTAGGTGGCCCACCG-C-CTCCACACA-----TGCCGCCA-----------CAC------A-----------------TGGCGCCCCAGCAGCAACAGCAGCAGCCG---CCTCAGCAG------CCACCA-CCTGGCGGC------ATG--CCTGGTCTGCCA-------------C--------------------------------CGCCGTTGCCCCACATGG-G--C-TATGCCA-------------ACTATGGTGGACCCGGTCCCGGCCAGCACTCGCACACGCACTACTACCAGCCGCAG---------------------------------------------------------TAT----GGACCGCACT---CGAG--A-------CCGCAG--CCCTACTACACGCACCTACCGCC | |
| droGri2 | scaffold_15110:6299479-6299708 - | CTGCCGC---CGC--------------------C--C-CCGT---CGCCG-CCAACGTC--------GC-------CC--ACGATCA------C------GC------------CTCCAATCACGCCCTGCAACCTCATCAGCAATAGCA--ACAAC--------------AACAACAATAGCAACAGCAA---CAGCAGCAACGGCAATAGCAACAACAACAACCA--ACAACAAC------AC------A--------------ATCA---TAACAA-------------------------------------------------------------------------------------------------------------------CATCAACAACAGTAACAA-----------CCACATTGACACGCCCAACA----------------A---CAGT---TTGCT---GC---TGAGCCC---CACTAAGCAGATGCT-------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/18/2015 at 04:17 AM