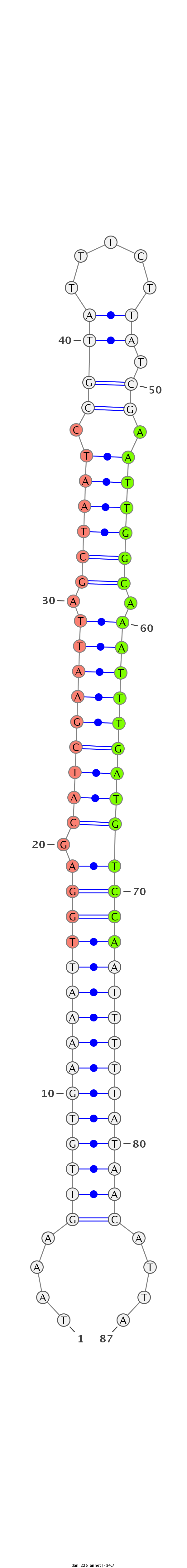
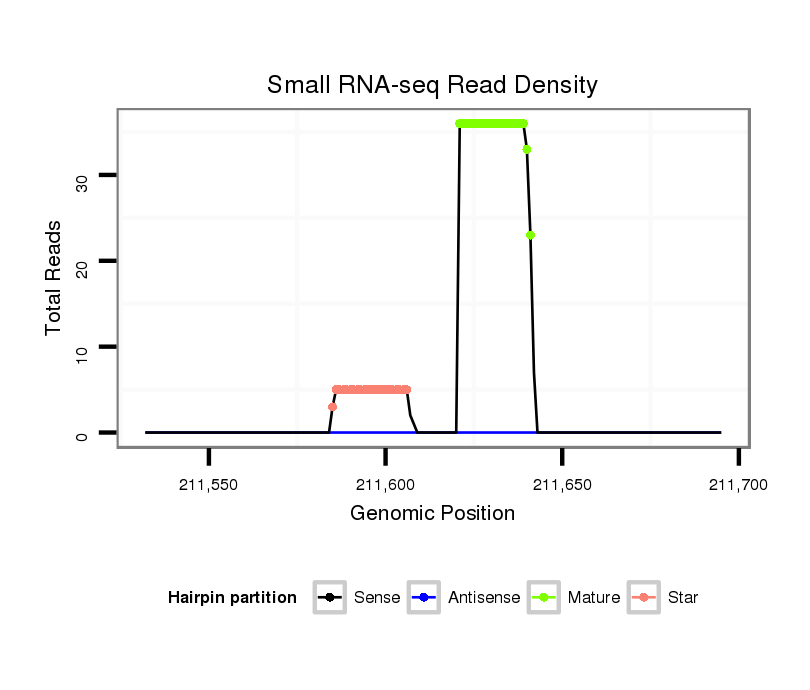
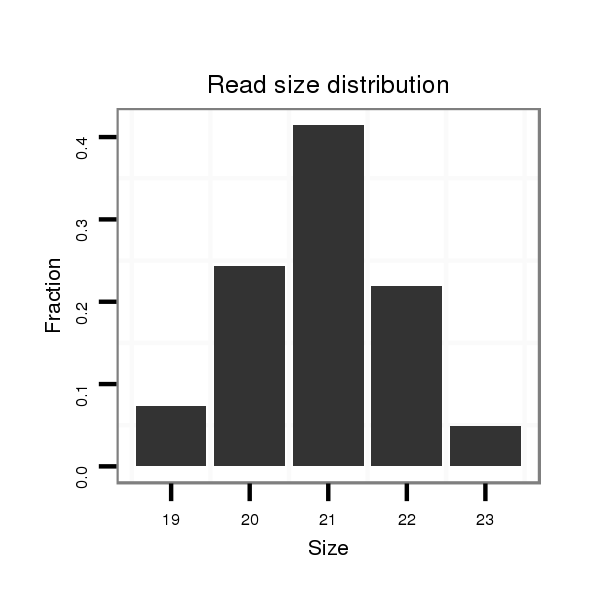

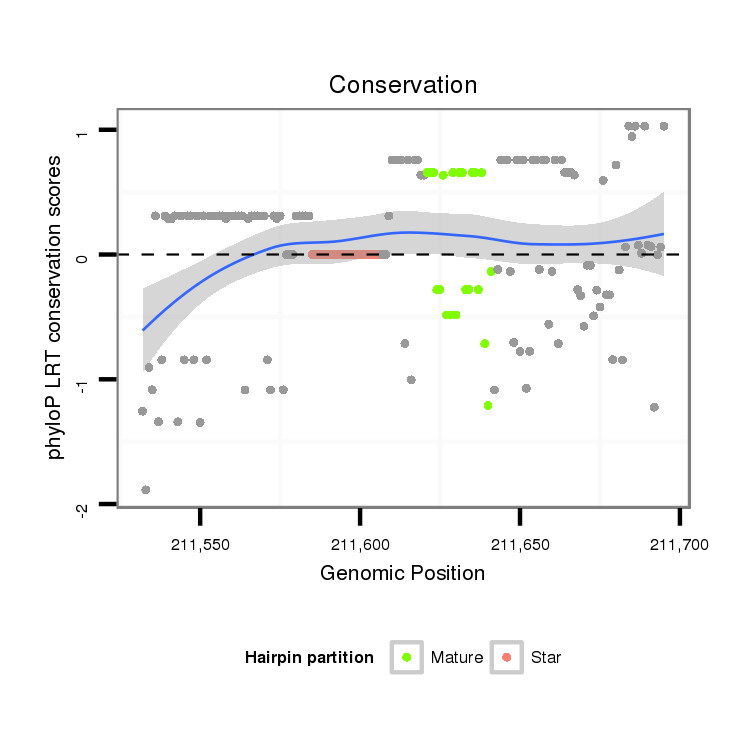
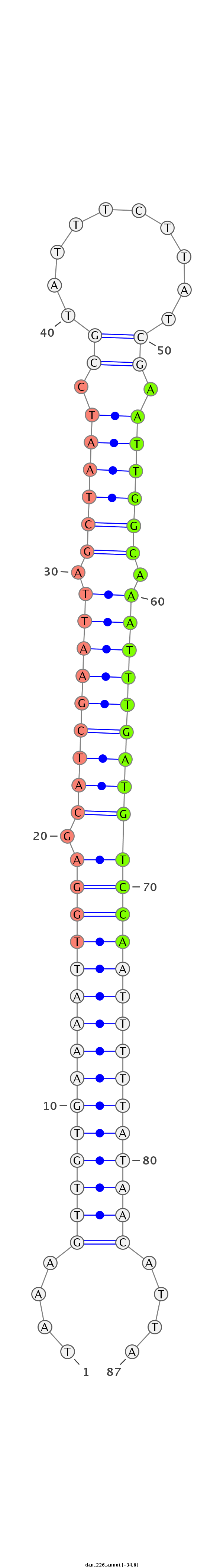
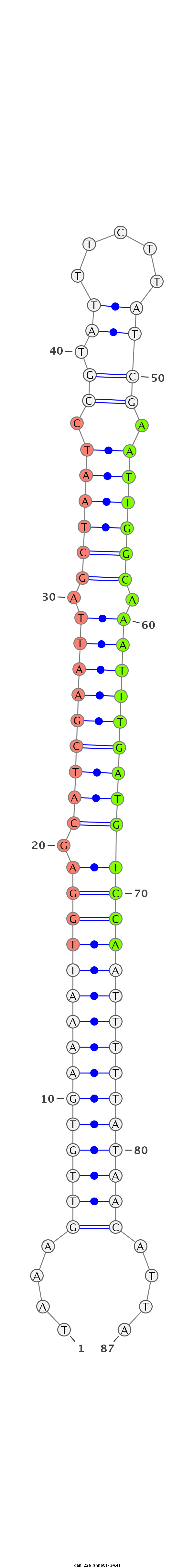
ID:dan_226 |
Coordinate:scaffold_13335:211582-211645 + |
Confidence:confident |
Class:Testes-restricted |
Genomic Locale:intergenic |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -34.6 | -34.4 | -34.1 |
 |
 |
 |
intergenic
No Repeatable elements found
|
ATTTTCGTCCTCAGATATGTAAAATTGTAAATCCTTTTTAAAGTTGTGAAAATTGGAGCATCGAATTAGCTAATCCGTATTTCTTATCGAATTGGCAAATTTGATGTCCAATTTTTATAACATTATTCATCTTAAGATTCGTGAGTGCTCTTATTTTAAACCTT
**************************************....(((((((((((((((.(((((((((.((((((.((((.....)).)).)))))).))))))))))))))))))))))))....*************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V105 male body |
V055 head |
V106 head |
M058 embryo |
|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................AATTGGCAAATTTGATGTCCA...................................................... | 21 | 0 | 1 | 16.00 | 16 | 5 | 7 | 4 | 0 |
| .........................................................................................AATTGGCAAATTTGATGTCC....................................................... | 20 | 0 | 1 | 10.00 | 10 | 6 | 1 | 1 | 2 |
| .........................................................................................AATTGGCAAATTTGATGTCCAA..................................................... | 22 | 0 | 1 | 7.00 | 7 | 2 | 3 | 2 | 0 |
| .........................................................................................AATTGGCAAATTTGATGTC........................................................ | 19 | 0 | 1 | 3.00 | 3 | 2 | 0 | 1 | 0 |
| .....................................................TGGAGCATCGAATTAGCTAATC......................................................................................... | 22 | 0 | 1 | 2.00 | 2 | 1 | 0 | 1 | 0 |
| .........................................................................................AATTGGCAGATTTGATGTCCA...................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 |
| ......................................................GGAGCATCGAATCAGCTAATCCG....................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ......................................................GGAGCATCGAATTAGCTAATCCG....................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .........................................................................................AATTGGCAGAGTTGATGTCCA...................................................... | 21 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ......................................................GGAGCATCGAATTAGCTAATC......................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .....................................................TGGAGCATCGAATTAGCTAATCC........................................................................................ | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| .........................................................................................AATTGTCAAATTTGATGTC........................................................ | 19 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 |
| .........................................................................................AATTGGTAAACTTGATGTC........................................................ | 19 | 2 | 4 | 0.50 | 2 | 2 | 0 | 0 | 0 |
|
TAAAAGCAGGAGTCTATACATTTTAACATTTAGGAAAAATTTCAACACTTTTAACCTCGTAGCTTAATCGATTAGGCATAAAGAATAGCTTAACCGTTTAAACTACAGGTTAAAAATATTGTAATAAGTAGAATTCTAAGCACTCACGAGAATAAAATTTGGAA
***************************************....(((((((((((((((.(((((((((.((((((.((((.....)).)).)))))).))))))))))))))))))))))))....************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V039 embryo |
V055 head |
|---|---|---|---|---|---|---|---|
| .............................................................................................................................ACGTAGAATTCTATGCACTCG.................. | 21 | 3 | 3 | 0.33 | 1 | 1 | 0 |
| ......................TGATCATTTAGGAGAAATTT.......................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 1 |
| Species | Coordinate | ID | Type | Alignment |
|---|---|---|---|---|
| droAna3 | scaffold_13335:211532-211695 + | dan_226 | confident | ATTT-------TCGTCCTCAGATATGTAAAATTGTAAATC---------------------CTTTTTAAAGTTGTGAAA---ATTGGAGCATCGAATTAGCTAATCCGTATTTCTTATCGAATTGGCAAATTTGATGTCCAATTTTTATAACATTATTCATCTTAAGATTCGTGAGTGCTCTTATTTTAAACCTT |
| droBip1 | scf7180000396778:4740-4871 - | TCCGTTGGGACTGATCCTGAAATGTCTGAAATTGTAAATATTGGTTTCCGACCATTTGTACCTTTTTGCAGTG---AAAACAAT------------------------TATTTTTGAT--------------------TAATTTTTTGTTATTTTATTTATTT---------------ATTTTA-TTCAAAACTT | ||
| droRho1 | scf7180000779514:101383-101412 + | AGA---------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTAAAGGCACTTGTATTTTAAACCTT | ||
| droBia1 | scf7180000302262:6636-6652 - | CTT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACATTTTAATTGCT | ||
| droEre2 | scaffold_4929:7279360-7279443 + | CAT----------------------------------------------------------------------------------------------------------ATTTTTTATCGAATCAGACACTTCAATATTTTGCTTTAATTAATTTGTTCTTTTTAAGGAGTATGA-----TTTATTATATATCTT |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 10/20/2015 at 06:51 PM