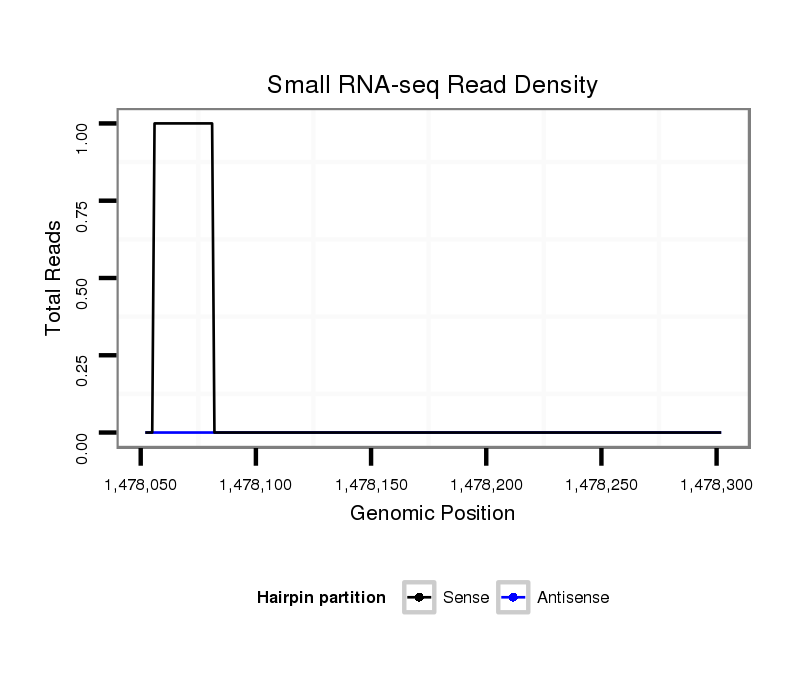
| CAACAA-CAGCA----------------------------------------------------------ACAACAACTGCAGCAGCAACAATTGCAG---CAACAACAGCAACAGCAGCAACAATGGCAGCAACAACAACAGCAACAACAACT---G----------------------------------------------------------------------CAGCAGCAA---CAACAACA-------------------------------------------------------------------------------GC-AGCAACAACAACTGCAGCAGCAACAATTGCAGCAA-C---AACAACAACAGCAGCAACAACAACAGCAGCAG------CAACAACAGCATCAG------------CAACAGC---A-ACAGCAGCAACGAATGCA | Size | Hit Count | Total Norm | Total | M020
Head | M045
Female-body | V117
Male-body | V118
Embryo | V119
Head |
| ......................................................................................................AACAACAGCAACAGCAGCAACAATGG................................................................................................................................................................................................................................................................................................................................................. | 26 | 20 | 0.35 | 7 | 0 | 7 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................................AATTGCAGCAA-C---AACAACAACAGC................................................................................... | 24 | 10 | 0.20 | 2 | 0 | 2 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................................................................................................................................................................CAA-C---AACAACAACAGCAGCA............................................................................... | 20 | 20 | 0.20 | 4 | 0 | 0 | 0 | 4 | 0 |
| .......................................................................................AACAATTGCAG---CAACAACAGCA................................................................................................................................................................................................................................................................................................................................................................. | 22 | 20 | 0.15 | 3 | 0 | 1 | 0 | 2 | 0 |
| .......................................................................................AACAATTGCAG---CAACAACAG................................................................................................................................................................................................................................................................................................................................................................... | 20 | 20 | 0.15 | 3 | 0 | 3 | 0 | 0 | 0 |
| ................................................................................................AG---CAACAACAGCAACAGCAGCAA....................................................................................................................................................................................................................................................................................................................................................... | 23 | 20 | 0.15 | 3 | 0 | 3 | 0 | 0 | 0 |
| ........................................................................................................................AACAATGGCAGCAACAACAACAGC................................................................................................................................................................................................................................................................................................................................. | 24 | 20 | 0.15 | 3 | 0 | 2 | 0 | 1 | 0 |
| ...................................................................................................................................................................................................................................................................................................................................................................................................................................TCAG------------CAACAGC---A-ACAGCAGCAACGAA.... | 26 | 9 | 0.11 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................................CAACAACTGCAGCAGCAACAATTGC......................................................................................................... | 25 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................GCAA---CAACAACA-------------------------------------------------------------------------------GC-AGCAACA................................................................................................................................ | 21 | 20 | 0.10 | 2 | 0 | 0 | 0 | 2 | 0 |
| ................................................................................................AG---CAACAACAGCAACAGCAG.......................................................................................................................................................................................................................................................................................................................................................... | 20 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................................................................................................ACAACAGCAGCAG------CAACAAC.................................................. | 20 | 20 | 0.10 | 2 | 0 | 0 | 0 | 0 | 2 |
| ...............................................................................................................................................................................................................................................................................................................................................................AACAATTGCAGCAA-C---AACAACAACAGC................................................................................... | 27 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................TGCAG---CAACAACAGCAACAGCAG.......................................................................................................................................................................................................................................................................................................................................................... | 23 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................................AATTGCAGCAA-C---AACAACAACAG.................................................................................... | 23 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................................................AGCAACAACAACAGCAACAACAACT---G----------------------------------------------------------------------............................................................................................................................................................................................................................................. | 26 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 |
| .......................................................................................................................CAACAATGGCAGCAACAACAACAG.................................................................................................................................................................................................................................................................................................................................. | 24 | 20 | 0.10 | 2 | 0 | 1 | 0 | 1 | 0 |
| .......................................................................CAACAACTGCAGCAGCAACAATTGC................................................................................................................................................................................................................................................................................................................................................................................. | 25 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................................................................................................................................AG------------CAACAGC---A-ACAGCAGCAACG...... | 22 | 11 | 0.09 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................................................................................................................CAACAGCATCAG------------CAACAGC---A-ACAGCAG........... | 27 | 12 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................................................................................CAACAGCAGCAG------CAACAACAGCATCAG------------.............................. | 27 | 12 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................................................................................CAACAACAGCATCAG------------CAACAGC---A-ACAG.............. | 27 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................................AATTGCAGCAA-C---AACAACA........................................................................................ | 19 | 13 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 |
| .........................................................................................................................................................T---G----------------------------------------------------------------------CAGCAGCAA---CAACAACA-------------------------------------------------------------------------------GC-AGCAACA................................................................................................................................ | 28 | 14 | 0.07 | 1 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................................................................................................................................................................................................................................................................................................................................................CAACAACAGCATCAG------------CAACAGC---.................... | 22 | 14 | 0.07 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...................................................................................................................................................AACAACT---G----------------------------------------------------------------------CAGCAGCAA---CAACAACA-------------------------------------------------------------------------------.......................................................................................................................................... | 25 | 15 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................................................................................................................AGCATCAG------------CAACAGC---A-ACAGCAG........... | 23 | 17 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................................................................................................................................................................................................CAACAGCATCAG------------CAACAGC---.................... | 19 | 17 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..................................................................................................................................................................................................................................................................................................................................................................................................................................ATCAG------------CAACAGC---A-ACAGCAGCAAC....... | 24 | 19 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................AGCAACAGCAGCAACAATGGCAG.............................................................................................................................................................................................................................................................................................................................................. | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................CAACAGCAACAGCAGCAACAATGG................................................................................................................................................................................................................................................................................................................................................. | 24 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................................................................................AATGGCAGCAACAACAACAGCAACAAC........................................................................................................................................................................................................................................................................................................................... | 27 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................................................................................................................CAACAACAACAGCAGCAG------C........................................................ | 19 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................................CAACAACTGCAGCAGCAACAATTGCAGC...................................................................................................... | 28 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................CAACAGCAACAACAACT---G----------------------------------------------------------------------CAG.......................................................................................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................ACAACAGCAACAGCAGCAACAATGG................................................................................................................................................................................................................................................................................................................................................. | 25 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................................................................................................................................................................AACAATTGCAGCAA-C---AACAACAG....................................................................................... | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................CAACAACAGCAACAACAACT---G----------------------------------------------------------------------CAGC......................................................................................................................................................................................................................................... | 25 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................AACAGCAACAACAACT---G----------------------------------------------------------------------CAGCAGCAA---C................................................................................................................................................................................................................................ | 27 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................................................................AACAACTGCAGCAGCAACAATTG.......................................................................................................... | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................TGGCAGCAACAACAACAGCAACAA............................................................................................................................................................................................................................................................................................................................ | 24 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| .AACAA-CAGCA----------------------------------------------------------ACAACAACTGCAG.............................................................................................................................................................................................................................................................................................................................................................................................. | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................................................AGCAACAACAACAGCAACAACAACT---G----------------------------------------------------------------------CAG.......................................................................................................................................................................................................................................... | 29 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................................................................................................................................................................................................................................AG------------CAACAGC---A-ACAGCAGCA......... | 19 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................AACAACTGCAGCAGCAACAATT................................................................................................................................................................................................................................................................................................................................................................................... | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................................................................................................................................................................................................................AACAACTGCAGCAGCAACAATT........................................................................................................... | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................CAACAATTGCAG---CAACAACAGCAA................................................................................................................................................................................................................................................................................................................................................................ | 24 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| .........................................................................................................................ACAATGGCAGCAACAACAACAGCA................................................................................................................................................................................................................................................................................................................................ | 24 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................................................................................................................................AACAACAACTGCAGCAGCAAC............................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| CAACAA-CAGCA----------------------------------------------------------ACAACAACTGCAGC............................................................................................................................................................................................................................................................................................................................................................................................. | 25 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................AACAACTGCAGCAGCAACAATTG.................................................................................................................................................................................................................................................................................................................................................................................. | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................ACAGCAACAACAACT---G----------------------------------------------------------------------CAGCA........................................................................................................................................................................................................................................ | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................AACAACAACT---G----------------------------------------------------------------------CAGCAGCAA---CA............................................................................................................................................................................................................................... | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................TGGCAGCAACAACAACAGCAACAACAA......................................................................................................................................................................................................................................................................................................................... | 27 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................................................................................AGCAACAACAACT---G----------------------------------------------------------------------CAGCAGCAA---................................................................................................................................................................................................................................. | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................AATTGCAG---CAACAACAGCAACAGC............................................................................................................................................................................................................................................................................................................................................................ | 24 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................AACAACAGCAACAGCAGCAACAATGGC................................................................................................................................................................................................................................................................................................................................................ | 27 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................AACAATTGCAG---CAACAACAGCAA................................................................................................................................................................................................................................................................................................................................................................ | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................................AGCAACAATGGCAGCAACAACAACA................................................................................................................................................................................................................................................................................................................................... | 25 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ....................................................................................................................................AACAACAACAGCAACAACAACT---G----------------------------------------------------------------------CAGC......................................................................................................................................................................................................................................... | 27 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................AATGGCAGCAACAACAACAGCAACA............................................................................................................................................................................................................................................................................................................................. | 25 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................................................................................................................................................................................ACAGCAGCAACAACAACAGCA................................................................... | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| ..............................................................................................................................GGCAGCAACAACAACAGCAACAA............................................................................................................................................................................................................................................................................................................................ | 23 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................................AA---CAACAACA-------------------------------------------------------------------------------GC-AGCAACAAC.............................................................................................................................. | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................AGCAACAGCAGCAACAATGG................................................................................................................................................................................................................................................................................................................................................. | 20 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................AACAATGGCAGCAACAACAACAGCA................................................................................................................................................................................................................................................................................................................................ | 25 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................................................................................AGCAACAACAACT---G----------------------------------------------------------------------CAGCAGCAA---CA............................................................................................................................................................................................................................... | 25 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ................................................................................................................................................AACAACAACT---G----------------------------------------------------------------------CAGCAGCAA---C................................................................................................................................................................................................................................ | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................AACAATTGCAG---CAACAACAGC.................................................................................................................................................................................................................................................................................................................................................................. | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........A----------------------------------------------------------ACAACAACTGCAGCAGCAACA...................................................................................................................................................................................................................................................................................................................................................................................... | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................................................................................................................................................................................................ACAACAGCAGCAACAACAACA...................................................................... | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................................................................TGGCAGCAACAACAACAGCAACAACAAC........................................................................................................................................................................................................................................................................................................................ | 28 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................AACAATTGCAG---CAACAACAGCAAC............................................................................................................................................................................................................................................................................................................................................................... | 24 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................ACAATGGCAGCAACAACAACAG.................................................................................................................................................................................................................................................................................................................................. | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................AACAACAACAGCAACAACAACT---G----------------------------------------------------------------------C............................................................................................................................................................................................................................................ | 24 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................................................................................................................................................................................AACAACAGCAGCAG------CAACAA................................................... | 20 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| .....................................................................................................CAACAACAGCAACAGCAGCAAC...................................................................................................................................................................................................................................................................................................................................................... | 22 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| .................................................................................................................CAGCAGCAACAATGGCAGCAA........................................................................................................................................................................................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................................TGCAG---CAACAACAGCAACAGCA........................................................................................................................................................................................................................................................................................................................................................... | 22 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .AACAA-CAGCA----------------------------------------------------------ACAACAACTGCAGCA............................................................................................................................................................................................................................................................................................................................................................................................ | 25 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................CAACAACTGCAGCAGCAACAATTGCAG---C........................................................................................................................................................................................................................................................................................................................................................................... | 28 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................AATGGCAGCAACAACAACAGC................................................................................................................................................................................................................................................................................................................................. | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................TTGCAG---CAACAACAGCAACAGCAG.......................................................................................................................................................................................................................................................................................................................................................... | 24 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ...........A----------------------------------------------------------ACAACAACTGCAGCAGCAAC....................................................................................................................................................................................................................................................................................................................................................................................... | 21 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................TTGCAG---CAACAACAGCAACAGCAGC......................................................................................................................................................................................................................................................................................................................................................... | 25 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |