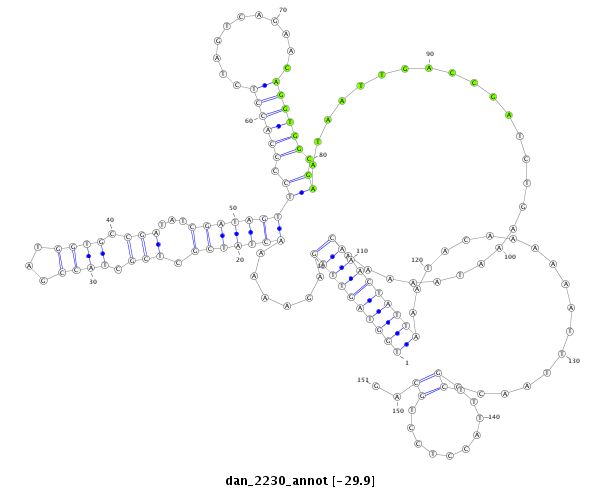
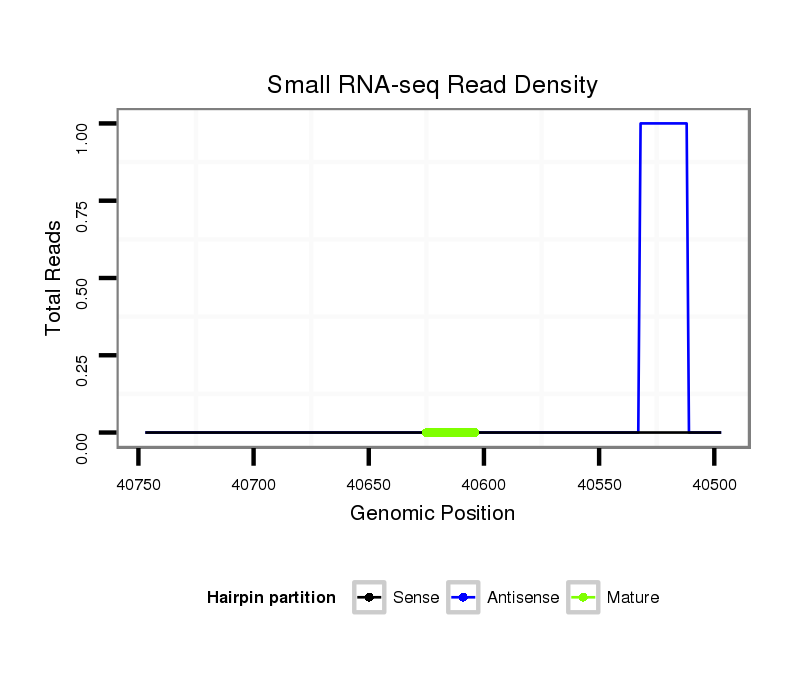
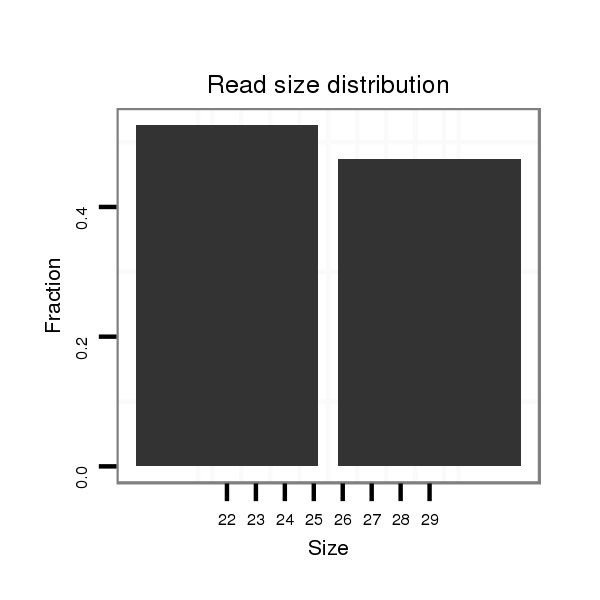
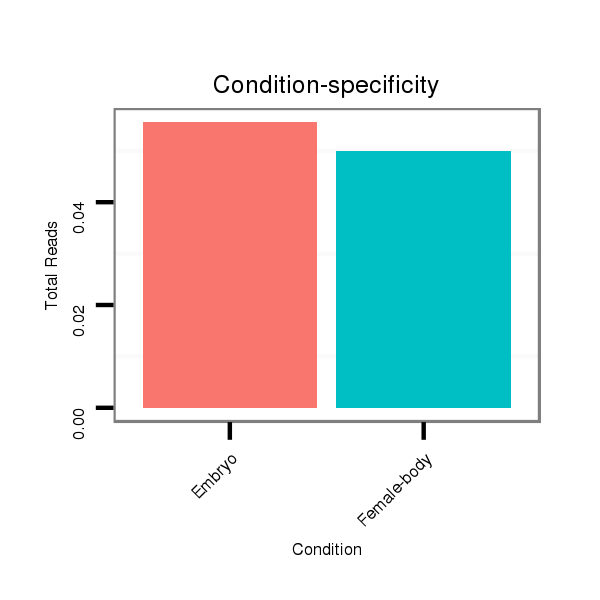
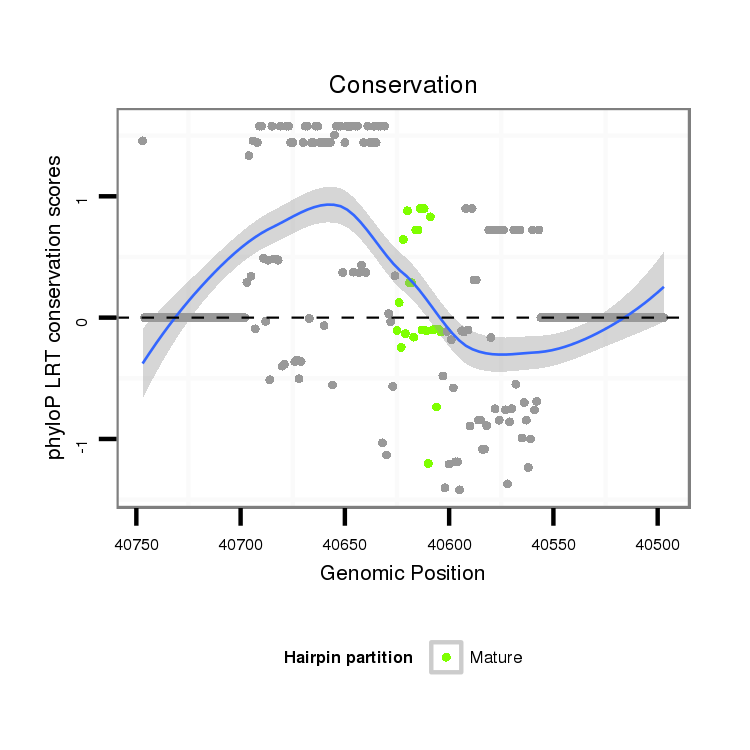
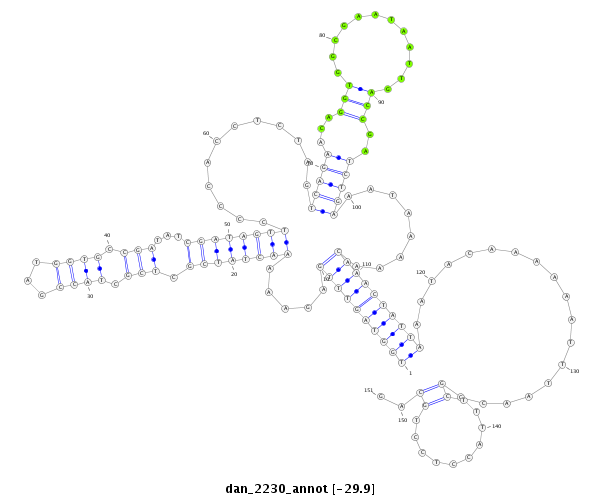
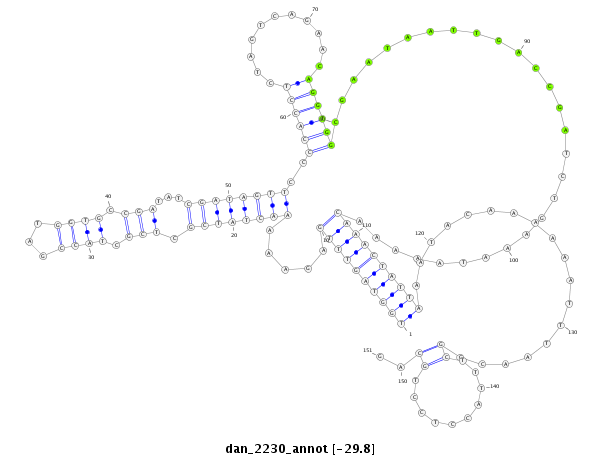
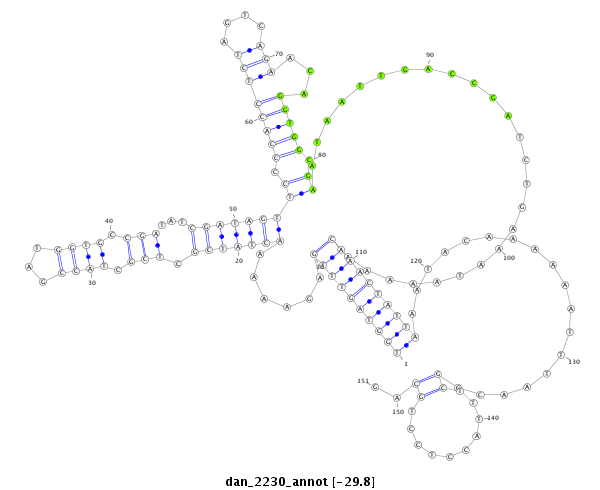
| .......................................................................................................................................................................................................................CCCACGCGGAGGAGGTCGTCA............... |
21 |
0 |
20 |
1.20 |
24 |
23 |
1 |
0 |
0 |
| ......................................................................................................................................................................................................................ACCCACGCGGAGGAGGTCGTC................ |
21 |
0 |
20 |
0.55 |
11 |
11 |
0 |
0 |
0 |
| ...............................................................................................................................................................................................................................GAGGAGGTCGTCACAGTCCAC....... |
21 |
0 |
20 |
0.40 |
8 |
6 |
2 |
0 |
0 |
| ..........................................................................................................GGTGGAGATCAGTCTTGTCCA............................................................................................................................ |
21 |
0 |
20 |
0.35 |
7 |
0 |
7 |
0 |
0 |
| ........................................................................CGAGCGATGGCTACCACGGCT.............................................................................................................................................................. |
21 |
0 |
20 |
0.35 |
7 |
7 |
0 |
0 |
0 |
| ..........................................CCCCTTGAACCATCAAACTCT............................................................................................................................................................................................ |
21 |
0 |
20 |
0.30 |
6 |
5 |
1 |
0 |
0 |
| ....................TCCGAATCGGTCATGATCTCC.................................................................................................................................................................................................................. |
21 |
0 |
20 |
0.25 |
5 |
5 |
0 |
0 |
0 |
| .....................CCGAATCGGTCATGATCTCC.................................................................................................................................................................................................................. |
20 |
0 |
20 |
0.20 |
4 |
4 |
0 |
0 |
0 |
| ...........CGGAATATATCCGAATCGGGC........................................................................................................................................................................................................................... |
21 |
1 |
20 |
0.20 |
4 |
4 |
0 |
0 |
0 |
| .......................................................................................................AGGGGTGGAGATCAGTCTTGT............................................................................................................................... |
21 |
0 |
20 |
0.15 |
3 |
3 |
0 |
0 |
0 |
| ...................................................................................................ATCAAGGGGTGGAGATCAGTC................................................................................................................................... |
21 |
0 |
20 |
0.15 |
3 |
3 |
0 |
0 |
0 |
| ..................................................................................................................................................................................................GGACGACACAGTCCCGTTATA.................................... |
21 |
3 |
7 |
0.14 |
1 |
0 |
0 |
1 |
0 |
| .............................GTCATGATCTCCGCCCCTTGA......................................................................................................................................................................................................... |
21 |
1 |
20 |
0.10 |
2 |
0 |
2 |
0 |
0 |
| ..............................................................................................................................................................................................................................GGAGGAGGTCGTCACAGTCC......... |
20 |
0 |
20 |
0.10 |
2 |
0 |
2 |
0 |
0 |
| ............................................................................................................TGGAGATCAGTCTTGTCCACC.......................................................................................................................... |
21 |
0 |
20 |
0.10 |
2 |
2 |
0 |
0 |
0 |
| ......................................................................................................AAGGGGTGGAGATCAGTCTTG................................................................................................................................ |
21 |
0 |
20 |
0.10 |
2 |
2 |
0 |
0 |
0 |
| ...........................CGGTCATGATCTCCACCCCTT........................................................................................................................................................................................................... |
21 |
0 |
20 |
0.10 |
2 |
2 |
0 |
0 |
0 |
| .........................................................................................................................................................................................................CCAGTCCCGTTTTACCCACGC............................. |
21 |
0 |
10 |
0.10 |
1 |
0 |
1 |
0 |
0 |
| ..........................................................................AGCGATGGCTACCACGGCTA............................................................................................................................................................. |
20 |
0 |
15 |
0.07 |
1 |
1 |
0 |
0 |
0 |
| ...................................................................................................................................................................................AATTGGCGAGACGGAGGA...................................................... |
18 |
2 |
16 |
0.06 |
1 |
0 |
0 |
1 |
0 |
| ....................................................................................................................................................................................................................TTCCCCACGCGGAGGAGGTCGT................. |
22 |
1 |
16 |
0.06 |
1 |
0 |
1 |
0 |
0 |
| .....................................................................................................................................................................................................CGTCGCAGTCCCGTTATA.................................... |
18 |
2 |
18 |
0.06 |
1 |
0 |
0 |
1 |
0 |
| .....................................................................................................................................................................................................................................GTCGTCACAGTCCACCCCAGC. |
21 |
0 |
20 |
0.05 |
1 |
0 |
1 |
0 |
0 |
| ......................CGAATCGGTCATGATCTCCAC................................................................................................................................................................................................................ |
21 |
0 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
| ............................................................................................................................................................................TTTTTTAAATTGGCGAGA............................................................. |
18 |
1 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
| ..................................................................TGATAGCGAGCGATGGCTACCACG................................................................................................................................................................. |
24 |
0 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
| ...................................................................................................................................................................................................................TTTCCCCACGCGGAGGAGGCC................... |
21 |
2 |
20 |
0.05 |
1 |
0 |
0 |
1 |
0 |
| ....................TCCGAATCGGTCATGGTCTCC.................................................................................................................................................................................................................. |
21 |
1 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
| ...........................................................................................................................................................................................................................CGCGGAGGAGGTCGTCACTG............ |
20 |
1 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
| ................................................................................................................................CGATCATTAACTGGCTAGA........................................................................................................ |
19 |
2 |
20 |
0.05 |
1 |
0 |
0 |
1 |
0 |
| ......................................................................................................................................................................................................................ACCCACGCGGAGGAGGTCCTC................ |
21 |
1 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
| ...........................................................CTCTTTTTGATAGCGAGCGATGGC........................................................................................................................................................................ |
24 |
0 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
| ................................................................TTTGATAGCGAGCGATGGCTA...................................................................................................................................................................... |
21 |
0 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
| ..............................................................................................................................................................................................................................GGAGGAGGTCGTCACAGTCCA........ |
21 |
0 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
| ........................................................................................................TGGGTGGAGATCAGTCTTGT............................................................................................................................... |
20 |
1 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
| .........................................................................................................GGGTGGAGATCAGTCTTGTCC............................................................................................................................. |
21 |
0 |
20 |
0.05 |
1 |
0 |
1 |
0 |
0 |
| ........................................................................................................................................................................................................................CCACGCGGAGGAGGTCGTCACA............. |
22 |
0 |
20 |
0.05 |
1 |
0 |
1 |
0 |
0 |
| ............................................................................................................................................................................................................................GCGGAGGAGGTCGTCACA............. |
18 |
0 |
20 |
0.05 |
1 |
0 |
1 |
0 |
0 |
| ............................GGTCATGATCTCCACCCCTTGAACCAT.................................................................................................................................................................................................... |
27 |
0 |
20 |
0.05 |
1 |
0 |
1 |
0 |
0 |
| ..............................................................................................................................................................................................................................GGAGGAGGTCGTCACTGTC.......... |
19 |
1 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
| .............................................................................................................................................................................................................................CGGAGGAGGTCGTCACAGTCC......... |
21 |
0 |
20 |
0.05 |
1 |
0 |
0 |
0 |
1 |