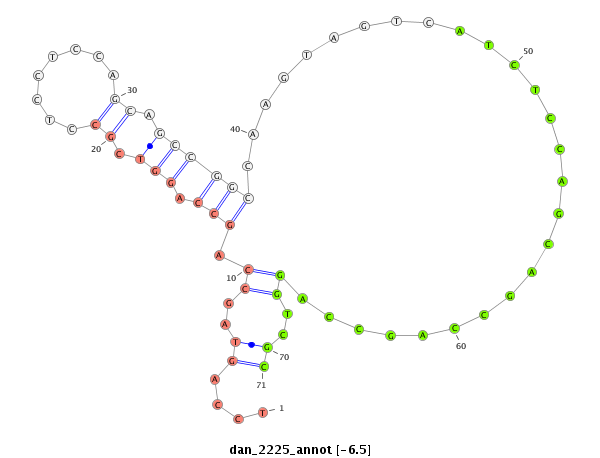
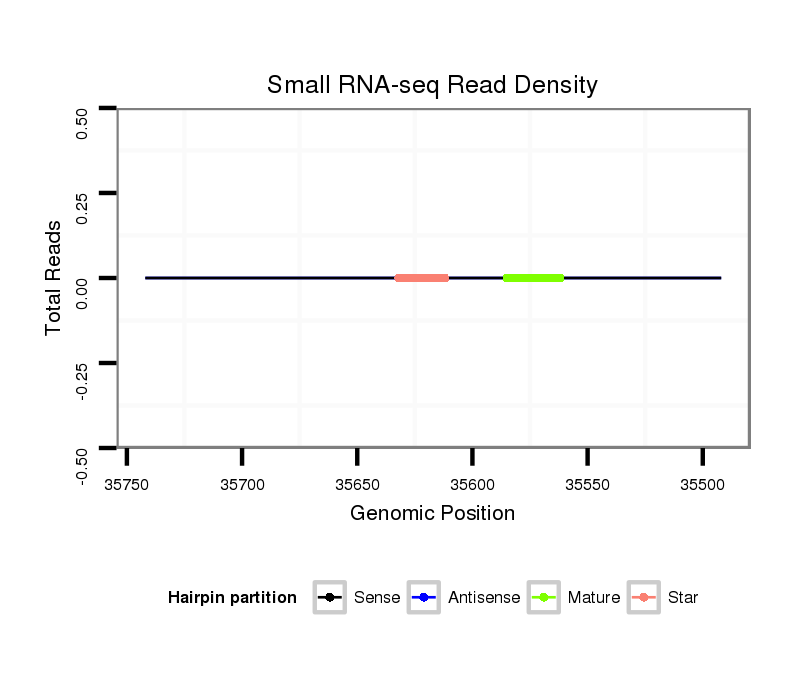
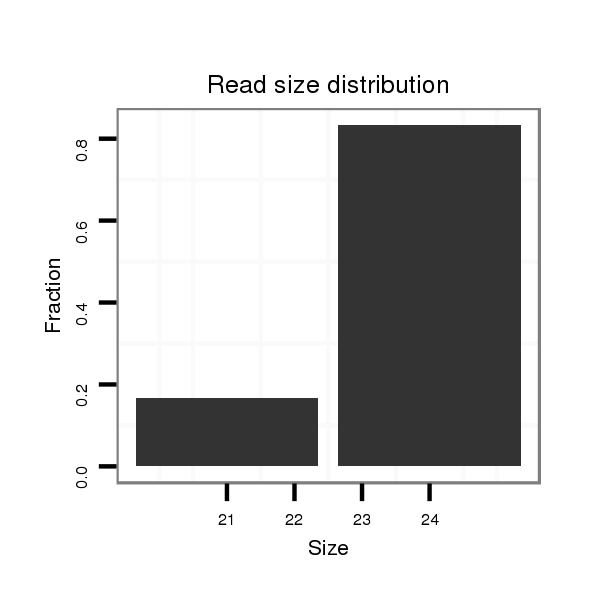
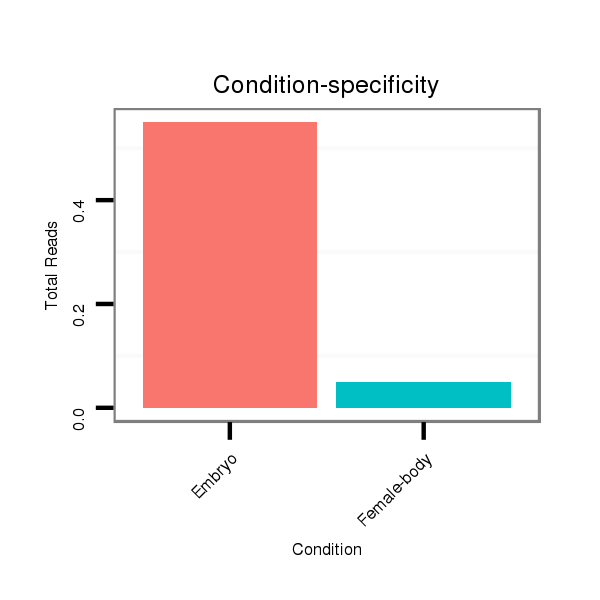
ID:dan_2225 |
Coordinate:scaffold_13150:35542-35692 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
No conservation details. |
| -14.4 |
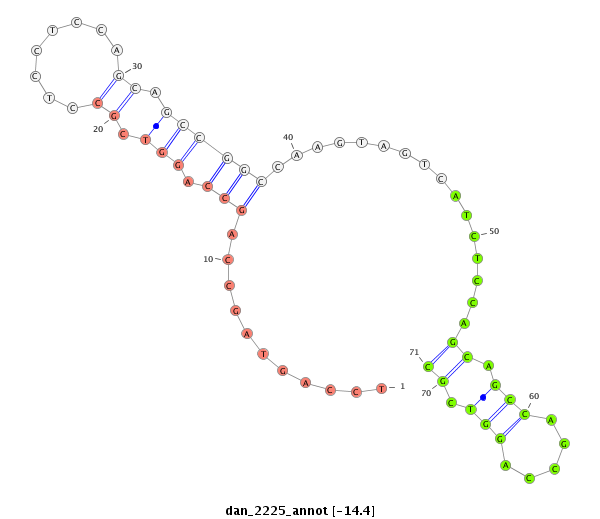 |
CDS [Dana\GF18978-cds]; exon [dana_GLEANR_20368:7]; intron [Dana\GF18978-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## CACCTCCTCCCGCAGCCAGCCAGGTCGCCACCTCCAGGTCAGTGGCATGGGCAGGGGTGTGCGCCTCCTCCAGCAGCCGCCCAGCTCGCCTCCTCCCGTCACCGGCCTCCTCCAGTAGCCAGCCAGGTCGCCTCCTCCAGCAGCCGGCCAAGTAGTCATCTCCAGCAGCCAGCCAGGTCGCCTCCTCCATTAGCCAGCCAGGTCGCCTCCACCAGGTCAGGGGCAGGGGCCAGAGAGGTCACCTCCTCCAG **************************************************************************************************************....((..((.(((.(((.((........)).))))))...........................))..))********************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M058 embryo |
M044 female body |
V039 embryo |
V055 head |
|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................................GATCTCCAGCAGCCAGCCAGGTCGC...................................................................... | 25 | 1 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................GCAGGGGCCAGAGAGGTCACC........ | 21 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................ATCTCCAGCAGCCAGCCAGGTCGC...................................................................... | 24 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................TCGCCTCCACCAGGTCAGGGG............................ | 21 | 0 | 20 | 0.25 | 5 | 5 | 0 | 0 | 0 |
| CACCTCCTCCCGCAGCCAGCCA..................................................................................................................................................................................................................................... | 22 | 0 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 |
| ............................................GCATGCGCAGCGGTGTTCGC........................................................................................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| CTCCTCCTCCCGCAGCCAGCCA..................................................................................................................................................................................................................................... | 22 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................CAGCCAGGTCGCCTCCTCCAT............................................................. | 21 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ..............................................................................................................TCCAGTAGCCAGCCAGGTCGC........................................................................................................................ | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................GACAGTGGCCAGAGAGGT............ | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
|
GTGGAGGAGGGCGTCGGTCGGTCCAGCGGTGGAGGTCCAGTCACCGTACCCGTCCCCACACGCGGAGGAGGTCGTCGGCGGGTCGAGCGGAGGAGGGCAGTGGCCGGAGGAGGTCATCGGTCGGTCCAGCGGAGGAGGTCGTCGGCCGGTTCATCAGTAGAGGTCGTCGGTCGGTCCAGCGGAGGAGGTAATCGGTCGGTCCAGCGGAGGTGGTCCAGTCCCCGTCCCCGGTCTCTCCAGTGGAGGAGGTC
**********************************************************************....((..((.(((.(((.((........)).))))))...........................))..))************************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M044 female body |
M058 embryo |
V055 head |
V106 head |
|---|---|---|---|---|---|---|---|---|---|
| .........................................................................TTCCGCGGGTCGAGCGGCGGAGGG.......................................................................................................................................................... | 24 | 3 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 |
| .......................................................................................................................................................................CGGTCGGTCCAGCGGAGGAGG............................................................... | 21 | 0 | 20 | 0.30 | 6 | 6 | 0 | 0 | 0 |
| .....................................................................................................................CGGTCGGTCCAGCGGAGGAGGT................................................................................................................ | 22 | 0 | 20 | 0.25 | 5 | 5 | 0 | 0 | 0 |
| ............GTCGGTCGGTCCAGCGGTGGA.......................................................................................................................................................................................................................... | 21 | 0 | 20 | 0.20 | 4 | 0 | 4 | 0 | 0 |
| ..............CGGTCGGTCCAGCGGTGGA.......................................................................................................................................................................................................................... | 19 | 0 | 20 | 0.15 | 3 | 0 | 3 | 0 | 0 |
| .......AGGGCGTCGGTCGGTCCAGC................................................................................................................................................................................................................................ | 20 | 0 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 |
| ...........................GGTGGAGGTCCAGTCACCGTA........................................................................................................................................................................................................... | 21 | 0 | 10 | 0.10 | 1 | 0 | 1 | 0 | 0 |
| .............................TGGAGGTCCAGTCACCGTGC.......................................................................................................................................................................................................... | 20 | 1 | 19 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................AGAGGTCGTCGGTCGGTCCAGCGG..................................................................... | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ........................................................................................................CGGAGGAGGTCATCGGTCGGTCCAGCG........................................................................................................................ | 27 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| .........................................................................................................................................................................GTCGGTCCAGCGGAGGAGGTA............................................................. | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ........................................TCATCGTAGCCGTCCACACA............................................................................................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| .......................................................................................................CCGGAGGAGGTCATCGTTCG................................................................................................................................ | 20 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ....................................................................................................................................................................................................CGGTCCAGCGGAGGTGGT..................................... | 18 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ...........CGTCGGTCGGTCCAGCGGTGGA.......................................................................................................................................................................................................................... | 22 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| .............TCGGTCGGTCCAGCGGTGGA.......................................................................................................................................................................................................................... | 20 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ....................................................................................GAGCGCAGGAGGGCGGTCG.................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| .......................................................................................................CCGGAGGAGGTCATCGGTCGGTCCAG.......................................................................................................................... | 26 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droAna3 | scaffold_13150:35492-35742 - | dan_2225 | CACCTCCTCCCGCAGCCAGCCAGGTCGCCACCTCCAGGTCAGTGGCATGGGCAGGGGTGTGCGCCTCCTCCAGCAGCCGCCCAGCTCGCCTCCTCCCGTCACCGGCCTCCTCCAGTAGCCAGCCAGGTCGCCTCCTCCAGCAGCCGGCCAAGTAGTCATCTCCAGCAGCCAGCCAGGTCGCCTCCTCCATTAGCCAGCCAGGTCGCCTCCACCAGGTCAGGGGCAGGGGCCAGAGAGGTCACCTCCTCCAG |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
Generated: 05/18/2015 at 01:49 AM