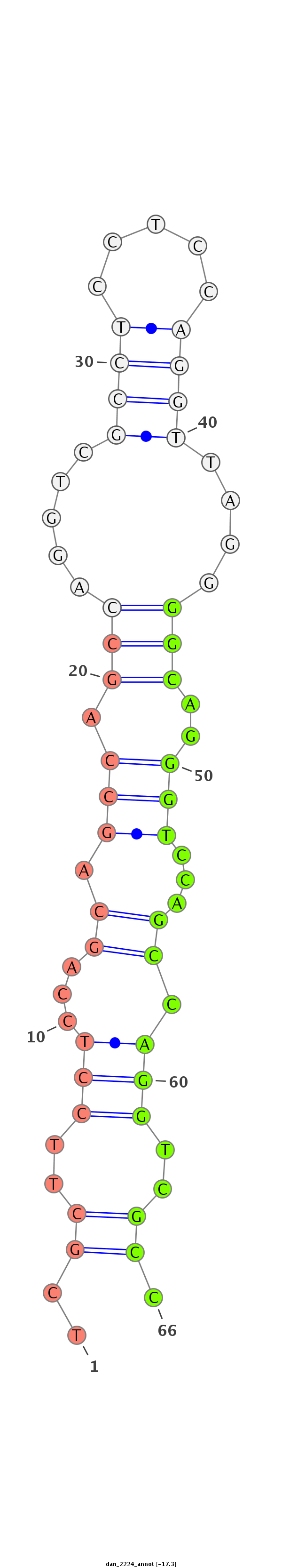
ID:dan_2224 |
Coordinate:scaffold_13150:34824-34974 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
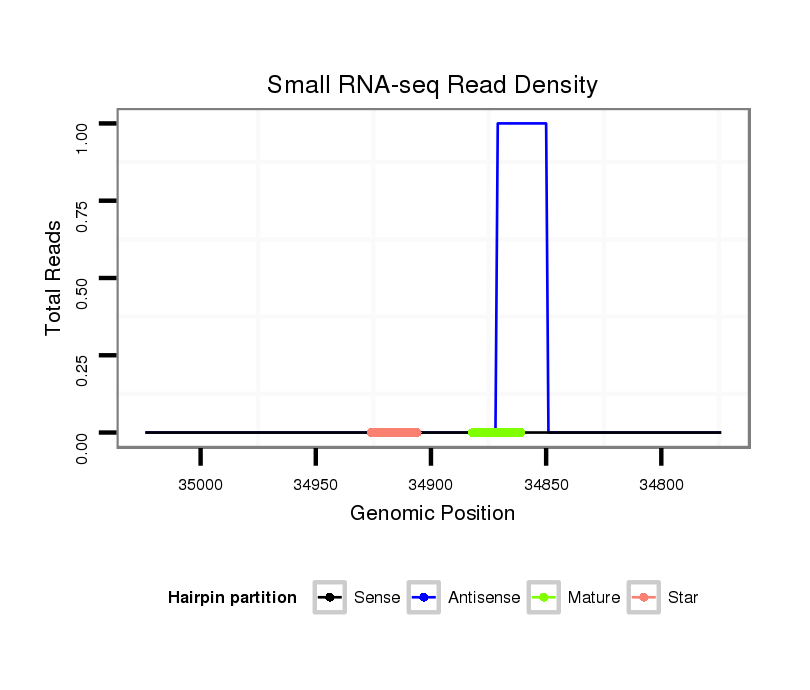
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
No conservation details. |
| -18.1 | -17.5 | -17.1 |
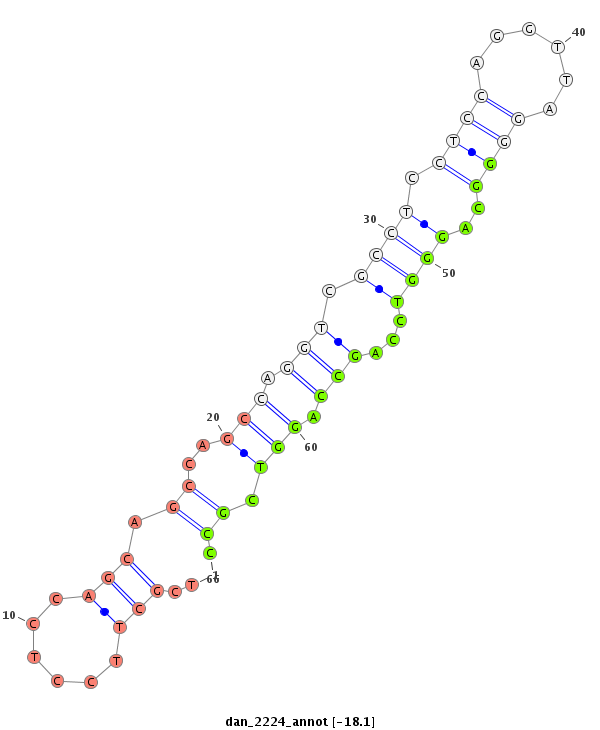 |
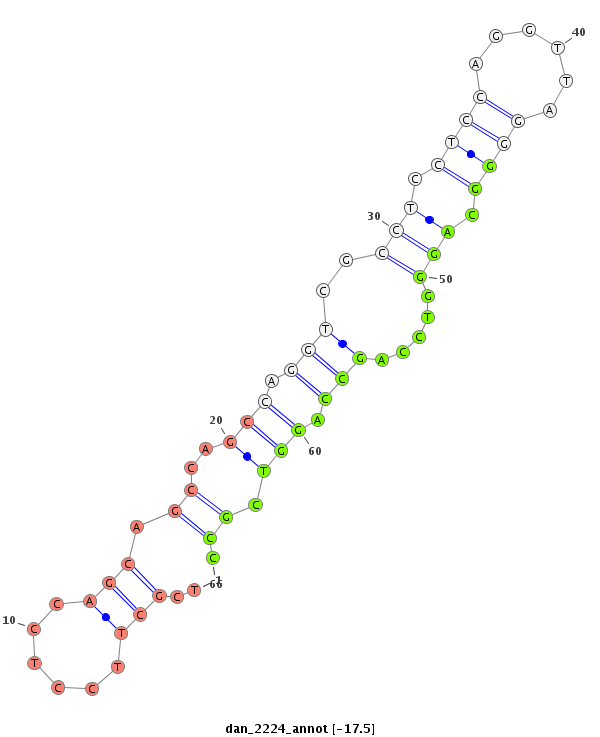 |
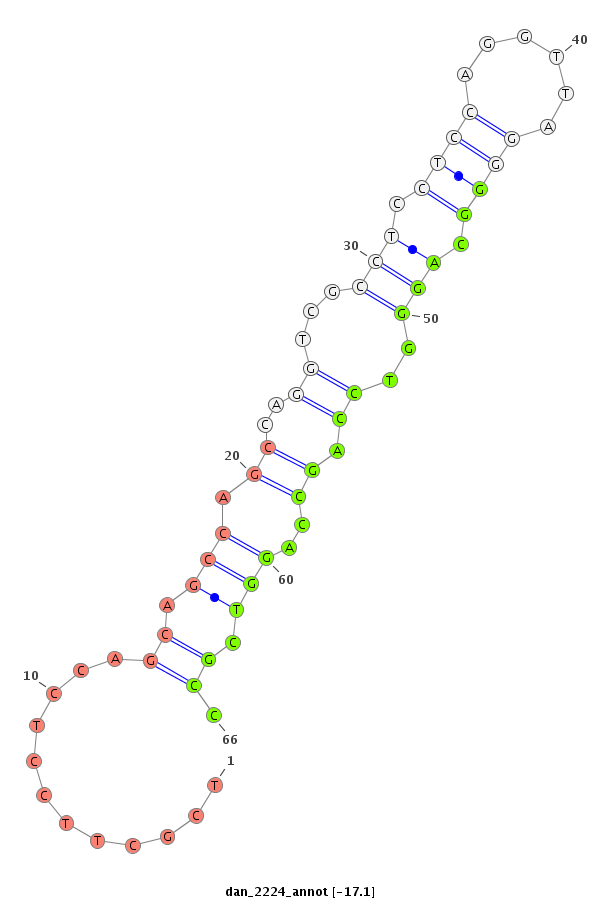 |
CDS [Dana\GF18978-cds]; exon [dana_GLEANR_20368:7]; intron [Dana\GF18978-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GGGCAGGGGTGTGCGCCTCCTCCAGTAGCCAGCAATGTCACCTCCTCCAGGTTAGGGGCAGGGGCCAGCCAGGTCGCCTCCTCCAGCTGCCAGCCAGGTCGCTTCCTCCAGCAGCCAGCCAGGTCGCCTCCTCCAGGTTAGGGGCAGGGTCCAGCCAGGTCGCCTCCTCCAGCAGCCAGCCAGGTCGCCTCCTCCAGGTCAGAGGCATGGGCAGGGGTGTGCGCCTCCTCCAGTAGCCAGCCAGGTCGCCT **************************************************************************************************..((..(((...((.(((.(((.....((((.....))))....)))..)))...)).)))..)).*************************************************************************************** |
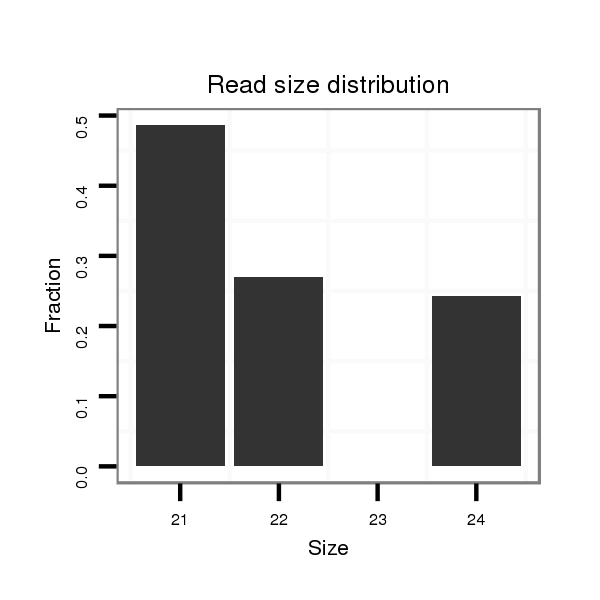
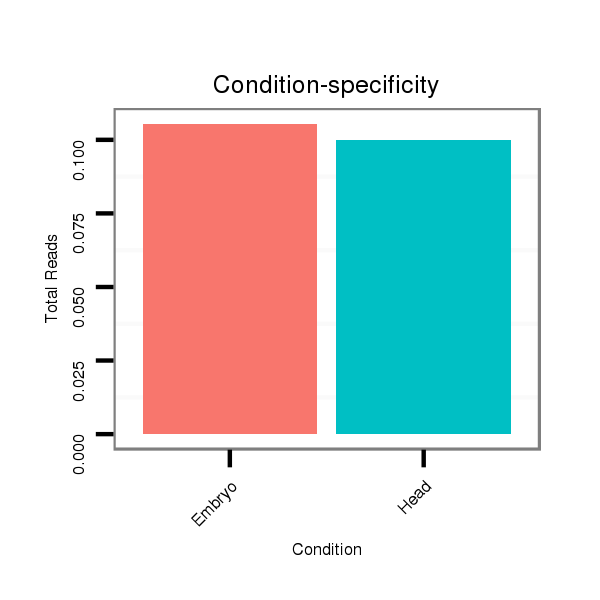
Read size | # Mismatch | Hit Count | Total Norm | Total | M058 embryo |
V106 head |
V039 embryo |
V055 head |
|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................GCTGCCAGCCAAGGCGCTTCC................................................................................................................................................. | 21 | 2 | 16 | 0.06 | 1 | 0 | 0 | 1 | 0 |
| ..............................................................................................................................................GGCAGGGTCCAGCCAGGTCGCC....................................................................................... | 22 | 0 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................GCATGCGCAGCGGTGTTCGC........................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| ...................................................................................................................................TCCAGGTTAGGGGCAGGGTCC................................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ..................................................................................................TCGCTTCCTCCAGCAGCCAGC.................................................................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ................................................................................................................................TCCTCCAGGTTAGGGGCAGGGTCC................................................................................................... | 24 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
|
CCCGTCCCCACACGCGGAGGAGGTCATCGGTCGTTACAGTGGAGGAGGTCCAATCCCCGTCCCCGGTCGGTCCAGCGGAGGAGGTCGACGGTCGGTCCAGCGAAGGAGGTCGTCGGTCGGTCCAGCGGAGGAGGTCCAATCCCCGTCCCAGGTCGGTCCAGCGGAGGAGGTCGTCGGTCGGTCCAGCGGAGGAGGTCCAGTCTCCGTACCCGTCCCCACACGCGGAGGAGGTCATCGGTCGGTCCAGCGGA
***************************************************************************************..((..(((...((.(((.(((.....((((.....))))....)))..)))...)).)))..)).************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M058 embryo |
M044 female body |
V039 embryo |
V055 head |
V106 head |
|---|---|---|---|---|---|---|---|---|---|---|
| .........................................................................................................................................................CGGTCCAGCGGAGGAGGTCGTC............................................................................ | 22 | 0 | 20 | 1.50 | 30 | 30 | 0 | 0 | 0 | 0 |
| ...................................................................................................................GTCGGTCCAGCGGAGGAGGT.................................................................................................................... | 20 | 0 | 20 | 0.45 | 9 | 0 | 9 | 0 | 0 | 0 |
| .............................................................................................................................................................CCAGCGGAGGAGGTCGTC............................................................................ | 18 | 0 | 20 | 0.40 | 8 | 8 | 0 | 0 | 0 | 0 |
| ..............................................................................................................CGTCGGTCGGTCCAGCGG........................................................................................................................... | 18 | 0 | 20 | 0.30 | 6 | 6 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................TCGGTCCAGCGGAGGAGGTC...................................................... | 20 | 0 | 20 | 0.25 | 5 | 5 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................GAGGTCGTCGGTCGGTCCAG................................................................. | 20 | 0 | 20 | 0.20 | 4 | 0 | 4 | 0 | 0 | 0 |
| ...............................................................................................................................................................................GGTCGGTCCAGCGGAGGAG......................................................... | 19 | 0 | 20 | 0.20 | 4 | 0 | 4 | 0 | 0 | 0 |
| .............................................................................................................................................CCCGTCCCAGGTCGGTCCAGC......................................................................................... | 21 | 0 | 18 | 0.17 | 3 | 3 | 0 | 0 | 0 | 0 |
| ......................................................................................................AAGGAGGTCGTCGGTCGGTCC................................................................................................................................ | 21 | 0 | 20 | 0.15 | 3 | 2 | 0 | 0 | 0 | 1 |
| .....................................................................................................................................................AGGTCGGTCCAGCGGAGGAGGT................................................................................ | 22 | 0 | 20 | 0.10 | 2 | 1 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................CCAGGTCGGTCCAGCGGAG..................................................................................... | 19 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................GGTCCAATCCCCGTCCCAGGT.................................................................................................. | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................CCGGTCGGTCCAGCGGAGGAGGTCG.................................................................................................................................................................... | 25 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................CTCCGAACCCGTCCCCGCGC.............................. | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................................................................................CCAGGTCGGTCCAGCGGAGGA................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................CCGTCCCAGGTCGGTCTATC......................................................................................... | 20 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................CGAAGGAGGTCGTCGGTCGGTCC................................................................................................................................ | 23 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| ..........CACGCGGAGGAGGTCATCGGT............................................................................................................................................................................................................................ | 21 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...............................................................................................................GTCGGTCGGTCCAGCGGAG......................................................................................................................... | 19 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..............................TCGTAACAGTGAAGGAGGA.......................................................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| .................................................................................................................................................CCCCAGGTCGGTCCAGCGGAG..................................................................................... | 21 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................CCAGCGGAGGAGGTCCAGT.................................................. | 19 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droAna3 | scaffold_13150:34774-35024 - | dan_2224 | GGGCAGGGGTGTGCGCCTCCTCCAGTAGCCAGCAATGTCACCTCCTCCAGGTTAGGGGCAGGGGCCAGCCAGGTCGCCTCCTCCAGCTGCCAGCCAGGTCGCTTCCTCCAGCAGCCAGCCAGGTCGCCTCCTCCAGGTTAGGGGCAGGGTCCAGCCAGGTCGCCTCCTCCAGCAGCCAGCCAGGTCGCCTCCTCCAGGTCAGAGGCATGGGCAGGGGTGTGCGCCTCCTCCAGTAGCCAGCCAGGTCGCCT |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
Generated: 05/18/2015 at 01:48 AM