ID:dan_2223 |
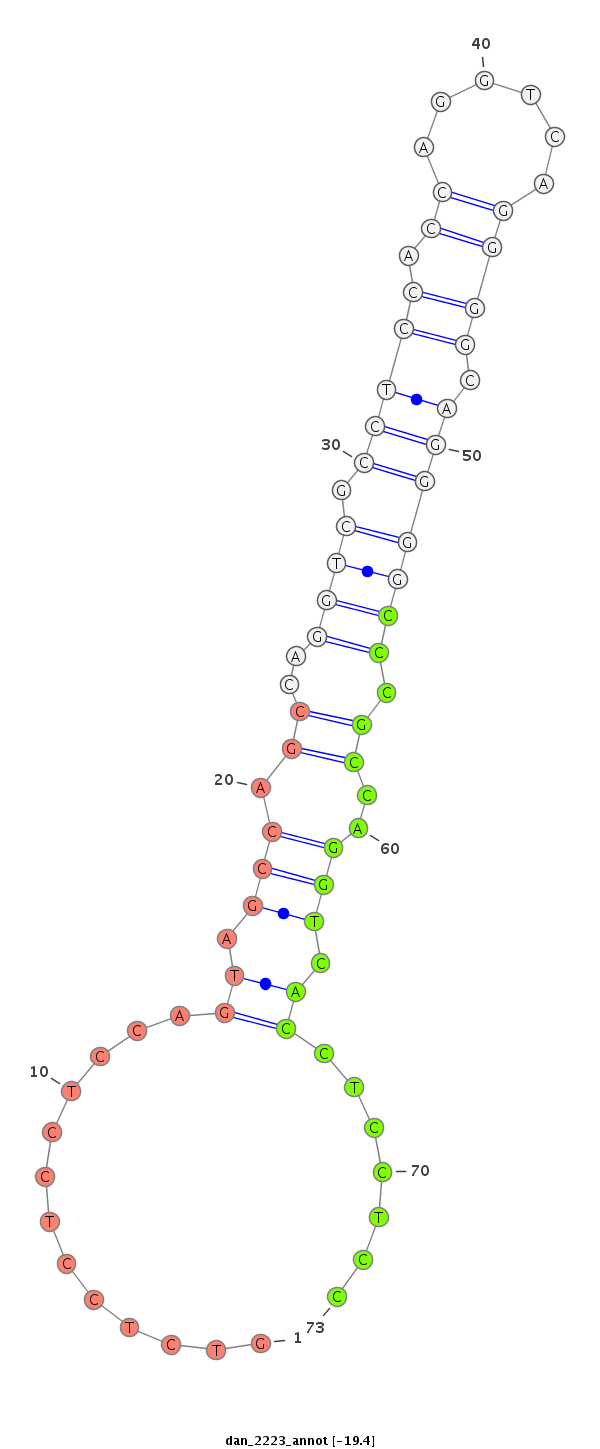
Coordinate:scaffold_13150:10209-10359 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
No conservation details. |
| -19.5 | -19.2 | -19.1 |
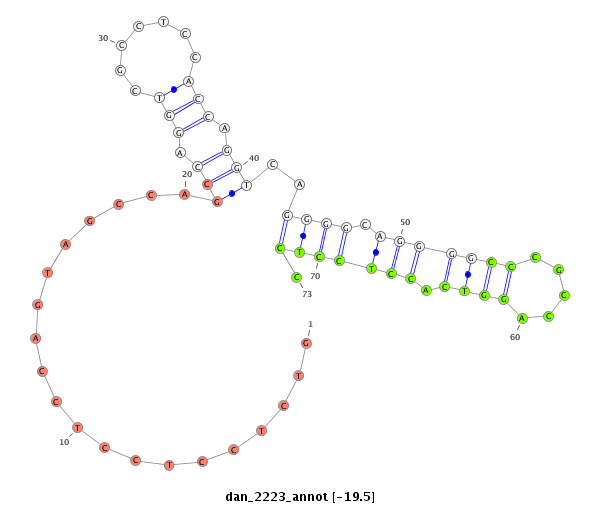 |
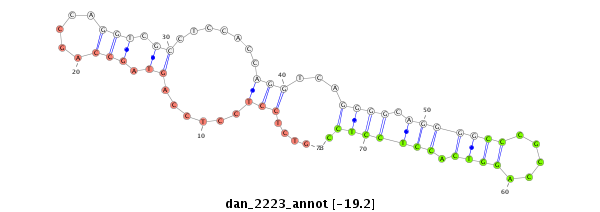 |
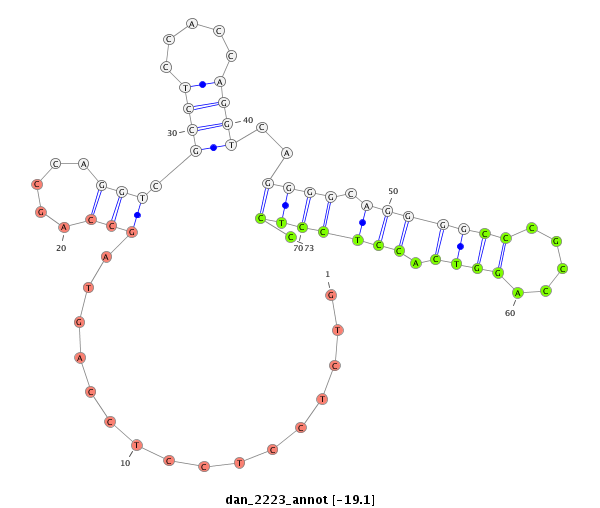 |
exon [dana_GLEANR_20368:8]; CDS [Dana\GF18978-cds]; intron [Dana\GF18978-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## AGCCGGCCAGGTCGCCTCCTTCAGCAGCCGGCCAGGTCGCTTCCTCCAGGTCAGTGGCGGGGCCAGGGGTGTGCGCCTCCTCCAGTAGCCAGCCAGGTCTCCTCCTCCAGTAGCCAGCCAGGTCGCCTCCACCAGGTCAGGGGCAGGGGCCCGCCAGGTCACCTCCTCCAGCAGCCAGCCAGGTCGCCTCCTCCAGGTTAGGGGCAGGAGCCAGCCAGGTGGGGTCGCCTCCTCCAGCAGCCGGCCAGGTC ************************************************************************************************.............((.(((.((..((((.(((((.((......)))).))))))).))..))).)).......********************************************************************************** |
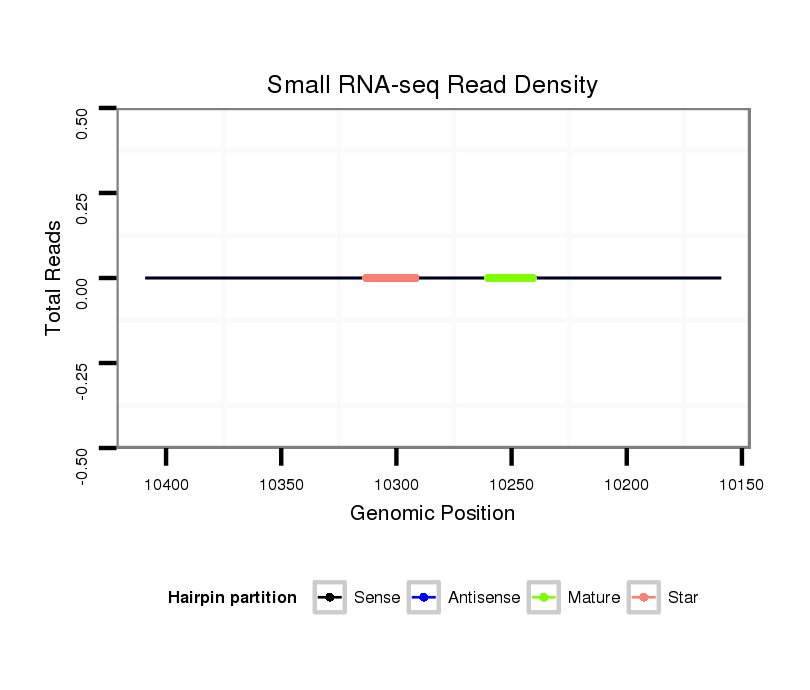
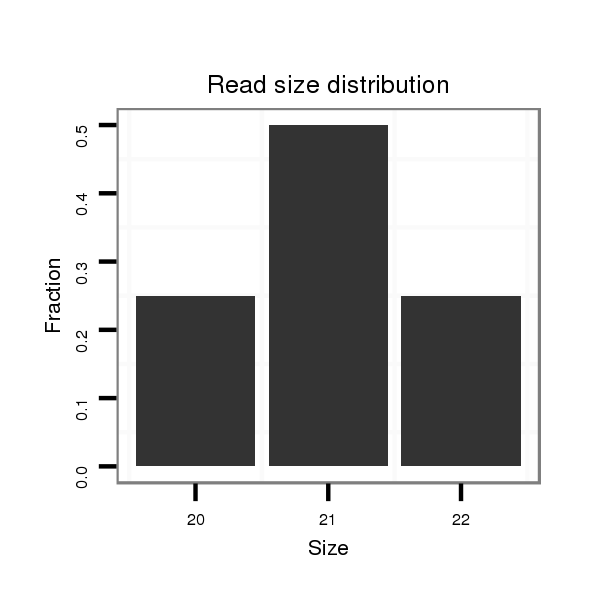
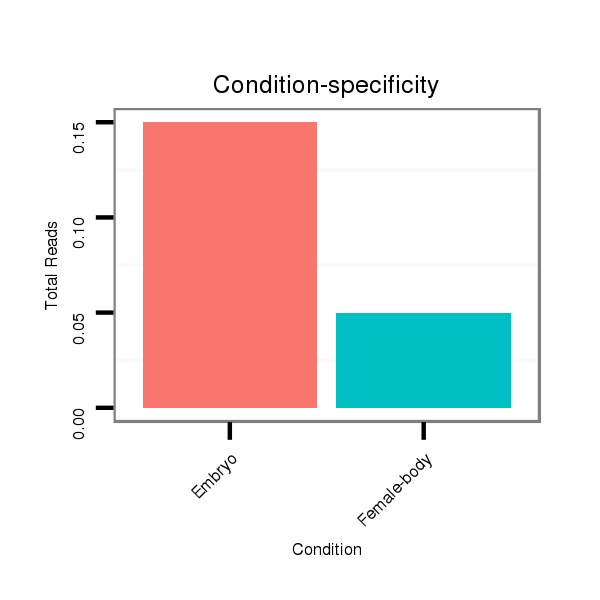
Read size | # Mismatch | Hit Count | Total Norm | Total | M058 embryo |
M044 female body |
|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................................................................................TCGCCTCCTCCAGCAGCCGGCC..... | 22 | 0 | 20 | 0.15 | 3 | 3 | 0 |
| ...........TCGCCTCCTTCAGCAGCCGGC........................................................................................................................................................................................................................... | 21 | 0 | 17 | 0.06 | 1 | 1 | 0 |
| .....................................................................................................................................................CCCGCCAGGTCACCTCCTCC.................................................................................. | 20 | 0 | 20 | 0.05 | 1 | 1 | 0 |
| ................................................................................................GTCTCCTCCTCCAGTAGCCAGC..................................................................................................................................... | 22 | 0 | 20 | 0.05 | 1 | 0 | 1 |
| .......................................................................................GCCAGCCAGGTCTCCTCCTCC............................................................................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 |
| ..................................GGTCGCTTCCTCCAGGTC....................................................................................................................................................................................................... | 18 | 0 | 20 | 0.05 | 1 | 1 | 0 |
| .........................................................................CGCCTCCTCCAGTAGCCAGCC............................................................................................................................................................. | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 |
|
TCGGCCGGTCCAGCGGAGGAAGTCGTCGGCCGGTCCAGCGAAGGAGGTCCAGTCACCGCCCCGGTCCCCACACGCGGAGGAGGTCATCGGTCGGTCCAGAGGAGGAGGTCATCGGTCGGTCCAGCGGAGGTGGTCCAGTCCCCGTCCCCGGGCGGTCCAGTGGAGGAGGTCGTCGGTCGGTCCAGCGGAGGAGGTCCAATCCCCGTCCTCGGTCGGTCCACCCCAGCGGAGGAGGTCGTCGGCCGGTCCAG
**********************************************************************************.............((.(((.((..((((.(((((.((......)))).))))))).))..))).)).......************************************************************************************************ |
Read size | # Mismatch | Hit Count | Total Norm | Total | M044 female body |
V039 embryo |
M058 embryo |
V105 male body |
|---|---|---|---|---|---|---|---|---|---|
| ................................................................................................................................................................TGGAGGAGGTCGTCGGTCGG....................................................................... | 20 | 0 | 20 | 0.20 | 4 | 4 | 0 | 0 | 0 |
| .......................................................................................................................................................................GGTCGTCGGTCGGTCCAGC................................................................. | 19 | 0 | 20 | 0.15 | 3 | 0 | 3 | 0 | 0 |
| ....CCGGTCCAGCGGAGGAAGTCG.................................................................................................................................................................................................................................. | 21 | 0 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 |
| ..............................................................................................................................................................................GGTCGGTCCAGCGGAGGAGGT........................................................ | 21 | 0 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 |
| .............................................................................................................................................................................................................................CCCAGCGGAGGAGGTCGTC........... | 19 | 0 | 20 | 0.10 | 2 | 0 | 0 | 2 | 0 |
| ..............................................................................................................................GAGGTGGTCCAGTCCCCGTCCC....................................................................................................... | 22 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| .............................................................................................................................................................................................................................CCCAGCGGAGGAGGTCGTCG.......... | 20 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| .............................................................................................................................................................................................................TCCTCGGTCGGACCACACCA.......................... | 20 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| ..........................................................................................................................................................................CGTCGGTCGGTCCAGCGGAGGAGGTC....................................................... | 26 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| ...........................................................................................CGGTCCAGAGGAGGAGGT.............................................................................................................................................. | 18 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ...............................GGGCCAGCGAAGGAGGTCCAGT...................................................................................................................................................................................................... | 22 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ................................................................................AGGTCATCGGTCGGTCCAGAG...................................................................................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................CCCAGCGGAGGAGGTCGTCGG......... | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| ......GGTCCAGCGGAGGAAGTCGTC................................................................................................................................................................................................................................ | 21 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| .............................................................................................................................................................................................................................CCCAGCGGAGGAGGTCGTCGGCCGGT.... | 26 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droAna3 | scaffold_13150:10159-10409 - | dan_2223 | AGCCGGCCAGGTCGCCTCCTTCAGCAGCCGGCCAGGTCGCTTCCTCCAGGTCAGTGGCGGGGCCAGGGGTGTGCGCCTCCTCCAGTAGCCAGCCAGGTCTCCTCCTCCAGTAGCCAGCCAGGTCGCCTCCACCAGGTCAGGGGCAGGGGCCCGCCAGGTCACCTCCTCCAGCAGCCAGCCAGGTCGCCTCCTCCAGGTTAGGGGCAGGAGCCAGCCAGGTGGGGTCGCCTCCTCCAGCAGCCGGCCAGGTC |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
Generated: 05/18/2015 at 01:48 AM