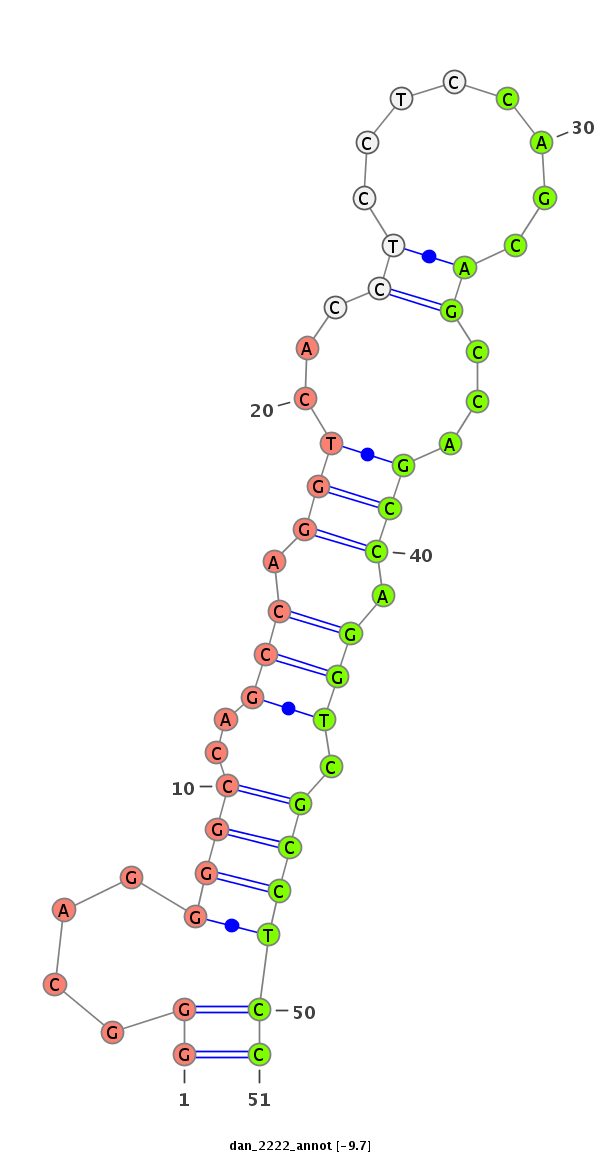
ID:dan_2222 |
Coordinate:scaffold_13150:6891-7041 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
No conservation details. |
| -17.2 | -16.6 |
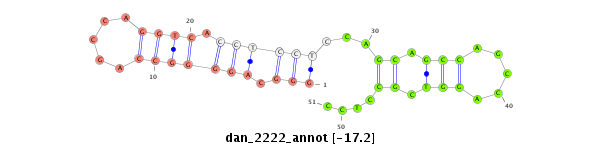 |
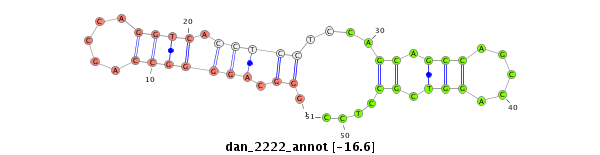 |
CDS [Dana\GF18978-cds]; exon [dana_GLEANR_20368:8]; intron [Dana\GF18978-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GTCGCCACCTCCAGCAGCCAGCCAGGTCGCCTCCTCCAGCAGCCGGCCAGGTCAGTGGTGGGGTCAGGGGTGTGCGCCTCCTCCAGTAGCCAGCCAGGTCGCCTCCTCCAGTAGCCAGCCAGGTCGCCTCCACCAGGTCAGGGGCAGGGGCCAGCCAGGTCACCTCCTCCAGCAGCCAGCCAGGTCGCCTCCTTCAGCAGCCAGCCAGGTCGCCTCCTACAGCAGCCGGCCAGGTCGCCTCCTCCAGGTCA *********************************************************************************************************************************************((....((((..(((.(((...((........))...))).))).))))))*********************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V106 head |
M058 embryo |
V055 head |
M044 female body |
|---|---|---|---|---|---|---|---|---|---|
| ........CTCCAGCAGCCAGCCAGGTCG.............................................................................................................................................................................................................................. | 21 | 0 | 20 | 0.45 | 9 | 9 | 0 | 0 | 0 |
| ....................................................CAGGGATGGGGTCCGGGGTGTG................................................................................................................................................................................. | 22 | 3 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 |
| ..............................................................................................................................................................................................................AGGTCGCCTCCTACCGCAGCC........................ | 21 | 1 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 |
| .........................................................................................................................................................................CAGCAGCCAGCCAGGTCGCCTCC........................................................... | 23 | 0 | 20 | 0.10 | 2 | 0 | 0 | 0 | 2 |
| .TCGCCACCTCCAGCAGCCAGCC.................................................................................................................................................................................................................................... | 22 | 0 | 14 | 0.07 | 1 | 0 | 1 | 0 | 0 |
| .........................................................GTGGGGTCTGGGGGGTGC................................................................................................................................................................................ | 18 | 2 | 15 | 0.07 | 1 | 0 | 0 | 1 | 0 |
| ................................................................................................................................................................................................................................GCCGGCCAGGTCGCCTCCTCC...... | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| ..................................................................................................................................................................................GCCAGGTCGCCTCCTTCAGCAGCCAGC.............................................. | 27 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| .............................................................................................................................................GGGCAGGGGCCAGCCAGGTCA......................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................ACAGCAGCTGGCCACGTC............... | 18 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ...........................................................................................................................................................................................................GCCAGATCGCCTCCTACAGCAGCC........................ | 24 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
|
CAGCGGTGGAGGTCGTCGGTCGGTCCAGCGGAGGAGGTCGTCGGCCGGTCCAGTCACCACCCCAGTCCCCACACGCGGAGGAGGTCATCGGTCGGTCCAGCGGAGGAGGTCATCGGTCGGTCCAGCGGAGGTGGTCCAGTCCCCGTCCCCGGTCGGTCCAGTGGAGGAGGTCGTCGGTCGGTCCAGCGGAGGAAGTCGTCGGTCGGTCCAGCGGAGGATGTCGTCGGCCGGTCCAGCGGAGGAGGTCCAGT
***********************************************************((....((((..(((.(((...((........))...))).))).))))))********************************************************************************************************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | M058 embryo |
V106 head |
M044 female body |
V039 embryo |
V105 male body |
|---|---|---|---|---|---|---|---|---|---|---|
| ...........................................................................................................................................................................................GGAGGAAGTCGTCGGTCGGTC........................................... | 21 | 0 | 20 | 0.20 | 4 | 0 | 0 | 4 | 0 | 0 |
| .......................................................................................................................................................................................CAGCGGAGGAAGTCGTCGGTC............................................... | 21 | 0 | 20 | 0.20 | 4 | 2 | 2 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................CGGTCCAGCGGAGGATGTCGTCG......................... | 23 | 0 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................CGGTCCAGCGGAGGATGTCGTC.......................... | 22 | 0 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................CGGTCCAGCGGAGGATGTCGTCGG........................ | 24 | 0 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................GGTCCAGCGGAGGATGTCGTC.......................... | 21 | 0 | 7 | 0.14 | 1 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................ATCGGTCGGTCCAGCGGAGGAGG.............................................................................................................................................. | 23 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................CGGTCGGTCCAGCGGAGGAAG........................................................ | 21 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 |
| ......TGGAGGTCGTCGGTCGGTCC................................................................................................................................................................................................................................. | 20 | 0 | 20 | 0.10 | 2 | 0 | 0 | 2 | 0 | 0 |
| .....................................................................................................................................................CGGTCGGTCCAGTGGAGG.................................................................................... | 18 | 0 | 20 | 0.10 | 2 | 0 | 0 | 2 | 0 | 0 |
| ........................................................................................................................................................................................................GGTCGGTCCAGCGGAGGATG............................... | 20 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 |
| ......TGGAGGTCGTCGGTCGGTCCAGC.............................................................................................................................................................................................................................. | 23 | 0 | 20 | 0.10 | 2 | 1 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................TTCGGTCCAGCGGAGGATGTCGTCG......................... | 25 | 1 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 |
| ......................................................CACCACCCGAGTCCACAC................................................................................................................................................................................... | 18 | 2 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................AGGTCGTCGGCCGGTCCAGTC.................................................................................................................................................................................................... | 21 | 0 | 17 | 0.06 | 1 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................................................................................................................................................................GTCCAGCGGAGGAGGTCCAGT | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| .....TTGGAGGTCGTCGGTCGGTC.................................................................................................................................................................................................................................. | 20 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| .....ATGGAGGTCGTCGGTCGGTCC................................................................................................................................................................................................................................. | 21 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................TCCAGCGGAGGAGGTCATCGGT...................................................................................................................................... | 22 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................CGGAGGAAGTCGTCGGTCGGTC........................................... | 22 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................TCGGTCCAGCGGAGGATG............................... | 18 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................CGGAGGAAGTCGTCGGTCGGTCCAGCG...................................... | 27 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...................................GGTCGTCGGCCGGTCCAGT..................................................................................................................................................................................................... | 19 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| .....TTGGAGGTCGTCGGTCGGTCC................................................................................................................................................................................................................................. | 21 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................CCACAGTTCCCACACGTGG............................................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...CGGTGGAGGTCGTCGGTCGG.................................................................................................................................................................................................................................... | 20 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................CGGTCCAGCGGAGGAGGTCGTCG................................................................................................................................................................................................................ | 23 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| .................................................................................................................................................................................TCGGTCCAGCGGAGGAAGT....................................................... | 19 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................ATCGGTCGGTCCAGCGGAGGA................................................................................................................................................ | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................GGAAGTCGTCGGTCGGTCCAGCGG..................................... | 24 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| .GGCGGTGGAGGTCGTCGGTC...................................................................................................................................................................................................................................... | 20 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droAna3 | scaffold_13150:6841-7091 - | dan_2222 | GTCGCCACCTCCAGCAGCCAGCCAGGTCGCCTCCTCCAGCAGCCGGCCAGGTCAGTGGTGGGGTCAGGGGTGTGCGCCTCCTCCAGTAGCCAGCCAGGTCGCCTCCTCCAGTAGCCAGCCAGGTCGCCTCCACCAGGTCAGGGGCAGGGGCCAGCCAGGTCACCTCCTCCAGCAGCCAGCCAGGTCGCCTCCTTCAGCAGCCAGCCAGGTCGCCTCCTACAGCAGCCGGCCAGGTCGCCTCCTCCAGGTCA |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
Generated: 05/18/2015 at 01:48 AM