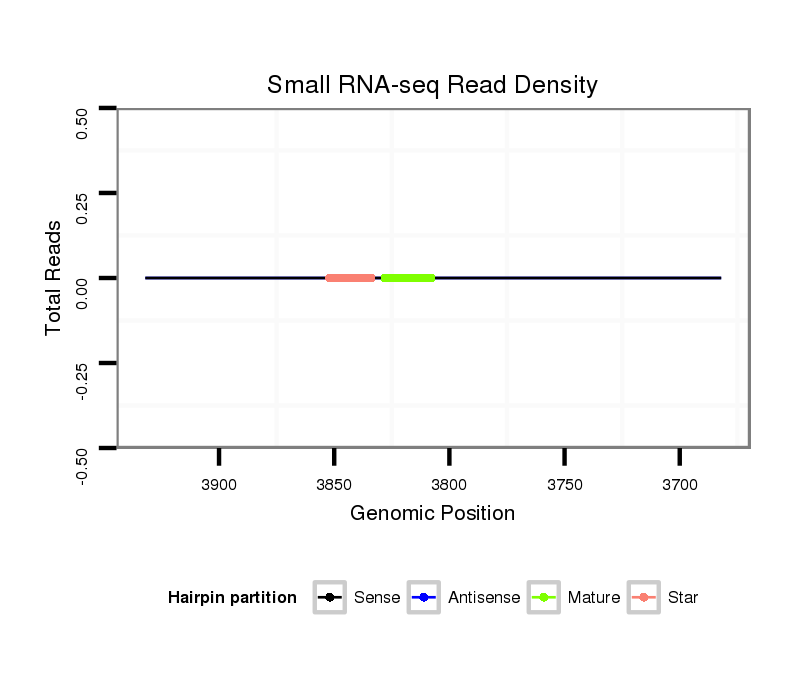
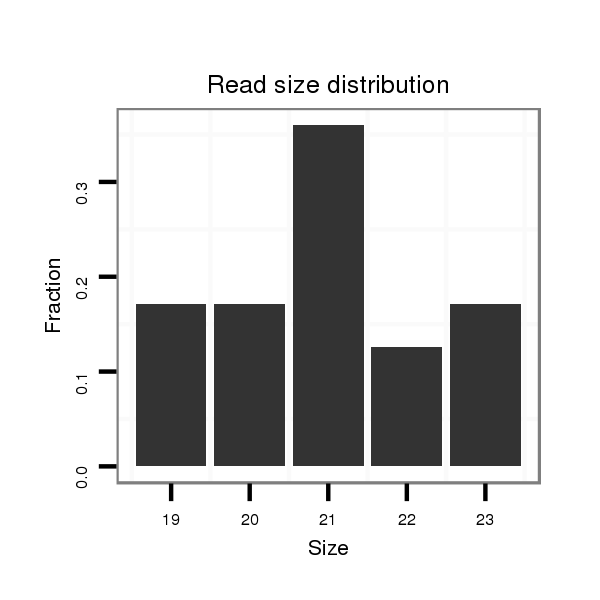
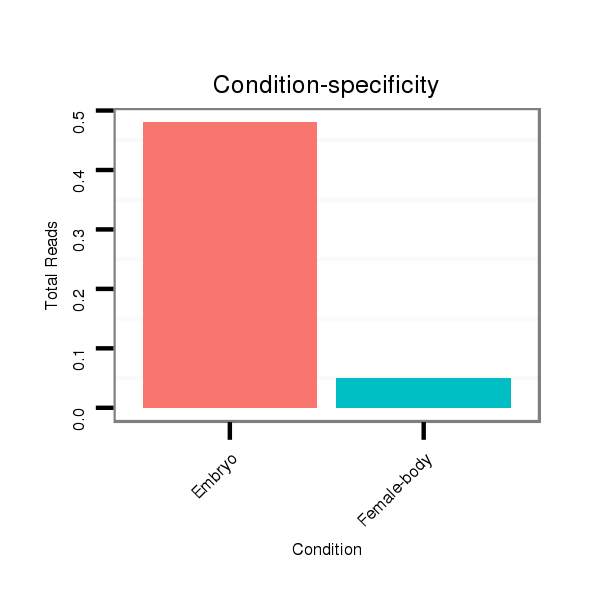
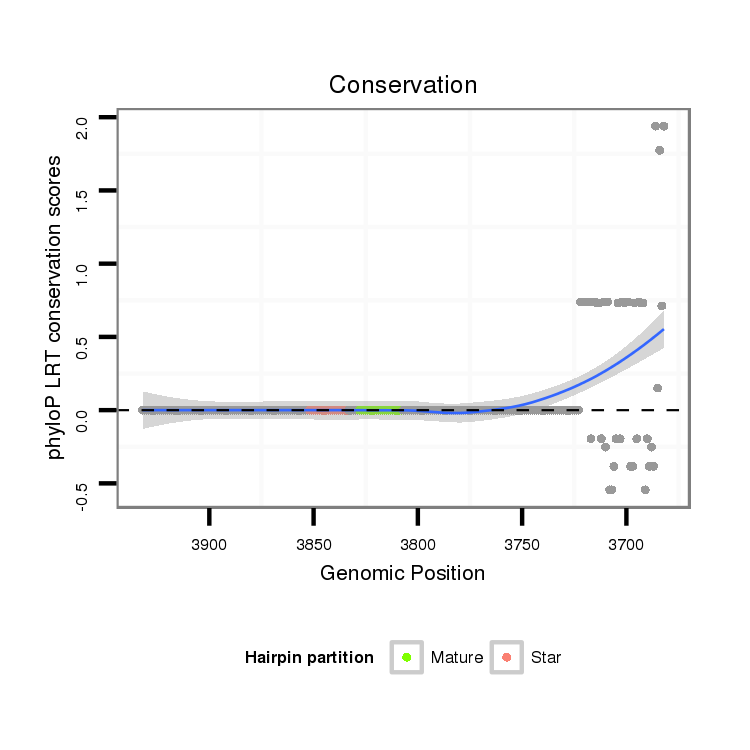
| .............................................................................TAGGCGGTCGTTGGTGGGTTC......................................................................................................................................................... |
21 |
3 |
12 |
0.42 |
5 |
0 |
5 |
0 |
0 |
0 |
| ............................................................CCCCGGTCGGTCCAGTGTA............................................................................................................................................................................ |
19 |
0 |
20 |
0.35 |
7 |
7 |
0 |
0 |
0 |
0 |
| ................................GGTCCAGCGGAGGAGGTCCAA...................................................................................................................................................................................................... |
21 |
0 |
20 |
0.35 |
7 |
0 |
0 |
0 |
7 |
0 |
| ..................................................................................................................................................................................................................................GTCATGATCTCCACCCCTTGA.... |
21 |
0 |
20 |
0.30 |
6 |
0 |
6 |
0 |
0 |
0 |
| ............................................................CCCCGGTCGGTCCAGTGTAG........................................................................................................................................................................... |
20 |
0 |
20 |
0.30 |
6 |
6 |
0 |
0 |
0 |
0 |
| .............................................................................TAGGCGGTCGTTCGTGGGTCC......................................................................................................................................................... |
21 |
3 |
9 |
0.22 |
2 |
0 |
1 |
0 |
0 |
1 |
| ..............................................................CCGGTCGGTCCAGTGTAGGCGG....................................................................................................................................................................... |
22 |
0 |
5 |
0.20 |
1 |
1 |
0 |
0 |
0 |
0 |
| .......CCTCCAGCGGAGGAGGTCGTCG.............................................................................................................................................................................................................................. |
22 |
0 |
5 |
0.20 |
1 |
0 |
0 |
1 |
0 |
0 |
| ........................................................CCGTCCCCGGTCGGTCCAGTGT............................................................................................................................................................................. |
22 |
0 |
20 |
0.20 |
4 |
4 |
0 |
0 |
0 |
0 |
| .......................................................................CCAGTGTAGGCGGTCGTCGGT............................................................................................................................................................... |
21 |
0 |
6 |
0.17 |
1 |
1 |
0 |
0 |
0 |
0 |
| .....................................................................GTCCAGTGTAGGCGGTCGTCGGTCG............................................................................................................................................................. |
25 |
0 |
6 |
0.17 |
1 |
0 |
0 |
1 |
0 |
0 |
| ..............................................GGTCCAATCCCCGTCCCCGGTC....................................................................................................................................................................................... |
22 |
0 |
20 |
0.15 |
3 |
3 |
0 |
0 |
0 |
0 |
| ...........................................................TCCCCGGTCGGTCCAGTGTAG........................................................................................................................................................................... |
21 |
0 |
20 |
0.15 |
3 |
0 |
3 |
0 |
0 |
0 |
| ..........................................................GTCCCCGGTCGGTCCAGTGTA............................................................................................................................................................................ |
21 |
0 |
20 |
0.15 |
3 |
0 |
3 |
0 |
0 |
0 |
| .............................................................................TAGGCGGTCGTCCGTGGGTT.......................................................................................................................................................... |
20 |
3 |
20 |
0.15 |
3 |
0 |
3 |
0 |
0 |
0 |
| .........................................................CGTCCCCGGTCGGTCCAGTGT............................................................................................................................................................................. |
21 |
0 |
20 |
0.15 |
3 |
0 |
3 |
0 |
0 |
0 |
| .......................................................................................................................................................................................................ACTTAAAGGGGAAATAGATA................................ |
20 |
2 |
7 |
0.14 |
1 |
0 |
1 |
0 |
0 |
0 |
| .................................................................................CGGTCGTCGGTCGGTCCAGCGG.................................................................................................................................................... |
22 |
0 |
15 |
0.13 |
2 |
2 |
0 |
0 |
0 |
0 |
| ................GAGGAGGTCGTCGGTAGGT........................................................................................................................................................................................................................ |
19 |
0 |
20 |
0.10 |
2 |
0 |
0 |
2 |
0 |
0 |
| ........................................................................CAGTGTTCGCGGTCCTCGGTC.............................................................................................................................................................. |
21 |
3 |
10 |
0.10 |
1 |
0 |
1 |
0 |
0 |
0 |
| ..............CGGAGGAGGTCGTCGGTAGG......................................................................................................................................................................................................................... |
20 |
0 |
20 |
0.10 |
2 |
0 |
0 |
2 |
0 |
0 |
| ......................................................CCCCGTCCCCGGTCGGTCCAGTGTA............................................................................................................................................................................ |
25 |
0 |
20 |
0.10 |
2 |
2 |
0 |
0 |
0 |
0 |
| ..............................................................CCGGTCGGTCCAGTGTAGG.......................................................................................................................................................................... |
19 |
0 |
20 |
0.10 |
2 |
2 |
0 |
0 |
0 |
0 |
| .............................................................CCCGGTCGGTCCAGTGTAGG.......................................................................................................................................................................... |
20 |
0 |
20 |
0.10 |
2 |
0 |
2 |
0 |
0 |
0 |
| ................................................................................GCGGTCGTCGGTCGGTCCAGC...................................................................................................................................................... |
21 |
0 |
11 |
0.09 |
1 |
0 |
0 |
0 |
1 |
0 |
| .............................................................................TAGGCGGTCGTCCGTGGGTTC......................................................................................................................................................... |
21 |
3 |
13 |
0.08 |
1 |
0 |
0 |
0 |
0 |
1 |
| .................................................................................CGGTCGTCGGTCGGTCCAG....................................................................................................................................................... |
19 |
0 |
16 |
0.06 |
1 |
0 |
0 |
1 |
0 |
0 |
| ..........................................................................................................................................AGGAAGTCACTGGCCAATA.............................................................................................. |
19 |
3 |
18 |
0.06 |
1 |
0 |
1 |
0 |
0 |
0 |
| ...........................CGGTAGGTCCAGCGGAGGAGG........................................................................................................................................................................................................... |
21 |
0 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
| ............................................................................GAAGGCGGTCGTCGGTCGGTC.......................................................................................................................................................... |
21 |
1 |
20 |
0.05 |
1 |
0 |
0 |
0 |
1 |
0 |
| ..........................TCGGTAGGTCCAGCGGAGGAGGT.......................................................................................................................................................................................................... |
23 |
0 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..........CCAGCGGAGGAGGTCGTCGGTAG.......................................................................................................................................................................................................................... |
23 |
0 |
20 |
0.05 |
1 |
0 |
0 |
1 |
0 |
0 |
| .........................GTCGGTAGGTCCAGCGGA................................................................................................................................................................................................................ |
18 |
0 |
20 |
0.05 |
1 |
0 |
0 |
1 |
0 |
0 |
| ...............................................................................GGCGTTCGTCGGTCGGTCC......................................................................................................................................................... |
19 |
1 |
20 |
0.05 |
1 |
0 |
0 |
0 |
1 |
0 |
| .........................................................CGTCCCCGGTCGGTCCAGTCT............................................................................................................................................................................. |
21 |
1 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
| .............GCGGAGGAGGTCGTCGGTAG.......................................................................................................................................................................................................................... |
20 |
0 |
20 |
0.05 |
1 |
0 |
0 |
1 |
0 |
0 |
| ......................................................CCCCGTCCCCGGTCGGTCCAGTCTAG........................................................................................................................................................................... |
26 |
1 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...................GAGGTCGTCGGTAGGTCCAGC................................................................................................................................................................................................................... |
21 |
0 |
20 |
0.05 |
1 |
0 |
1 |
0 |
0 |
0 |