
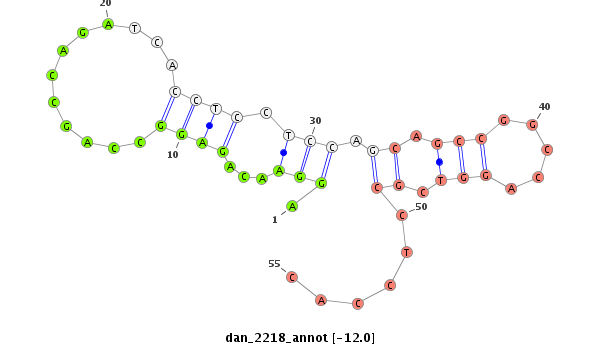
ID:dan_2218 |
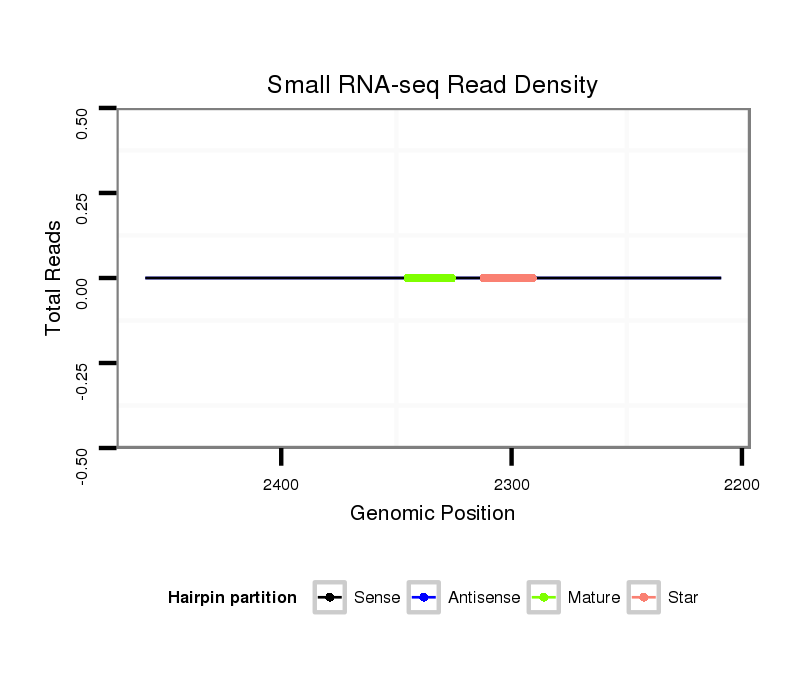
Coordinate:scaffold_13150:2259-2409 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
No conservation details. |
| -12.0 |
 |
exon [dana_GLEANR_20368:10]; CDS [Dana\GF18978-cds]; intron [Dana\GF18978-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GGCCAGGGGGTTGCGCCCCCTCCAGCAGCCGGCCAGGTCGACTCCTCCAGGTTAGGGGCAGCAGCCAGCCAGGTCACCTCCTCCAGCAGCCAGCCAGATCGTCCCCTCCAGGTTAGGAACAGAGGCCAGCCAGATCACCTCCTCCAGCAGCCGGCCAGGTCGCCTCCACAAGNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN ******************************************************************************************************************.((.......(((.(((.....................)))..))).....))..********************************************************************************** |
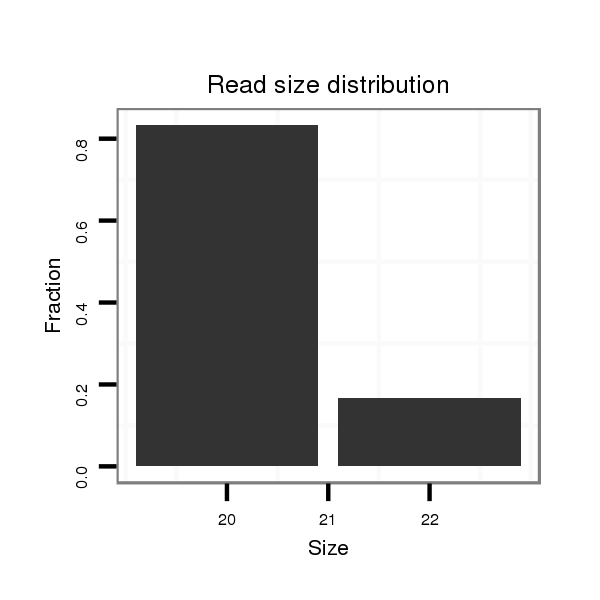
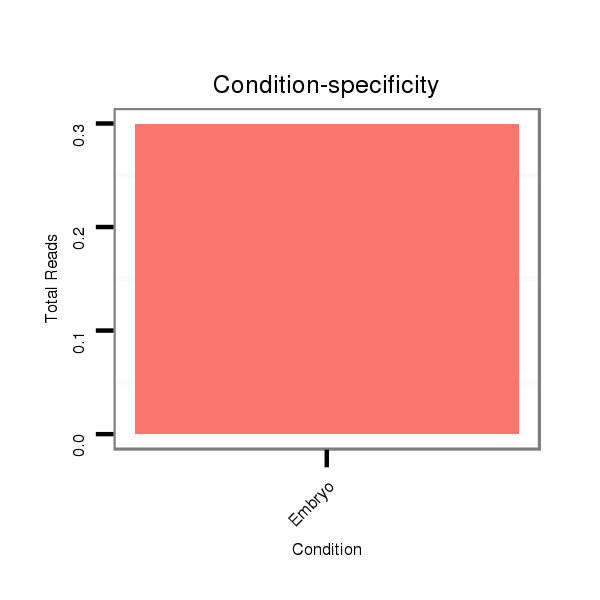
Read size | # Mismatch | Hit Count | Total Norm | Total | M058 embryo |
|---|---|---|---|---|---|---|
| ..................................................................................................................AGGAACAGAGGCCAGCCAGA..................................................................................................................... | 20 | 0 | 4 | 0.25 | 1 | 1 |
| ...................................................................................................................................................CAGCCGGCCAGGTCGCCTCCAC.................................................................................. | 22 | 0 | 20 | 0.05 | 1 | 1 |
|
CCGGTCCCCCAACGCGGGGGAGGTCGTCGGCCGGTCCAGCTGAGGAGGTCCAATCCCCGTCGTCGGTCGGTCCAGTGGAGGAGGTCGTCGGTCGGTCTAGCAGGGGAGGTCCAATCCTTGTCTCCGGTCGGTCTAGTGGAGGAGGTCGTCGGCCGGTCCAGCGGAGGTGTTCNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN
**********************************************************************************.((.......(((.(((.....................)))..))).....))..****************************************************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M058 embryo |
M044 female body |
V106 head |
|---|---|---|---|---|---|---|---|---|
| ........................................................CCGTCGTCGGTCGGTCCAGTG.............................................................................................................................................................................. | 21 | 0 | 20 | 0.55 | 11 | 11 | 0 | 0 |
| ................................................................................................CTAGCAGGGGAGGTCCAATC....................................................................................................................................... | 20 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 |
| ...........................................................................TGGAGGAGGTCGTCGGTCGGT........................................................................................................................................................... | 21 | 0 | 20 | 0.10 | 2 | 0 | 0 | 2 |
| ........................CGTCGGCCGGTCCAGCTG................................................................................................................................................................................................................. | 18 | 0 | 15 | 0.07 | 1 | 1 | 0 | 0 |
| ............................GGCCGGTCCAGCTGAGGAGG........................................................................................................................................................................................................... | 20 | 0 | 19 | 0.05 | 1 | 0 | 1 | 0 |
| ........................................TGAGGAGGTCCAATCCCCGTC.............................................................................................................................................................................................. | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| ....................................................................................................................................CTAGTGGAGGAGGTCGTC..................................................................................................... | 18 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| ............................................................................................................................CGGTCGGTCTAGTGGAGGATG.......................................................................................................... | 21 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| ......................................................................................................................................................GGCCGGTCGAGCGGAGGTGTT................................................................................ | 21 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| .........................................................................................................................................................CGGTCCAGTGGAGGTGTT................................................................................ | 18 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| .............................................................................GAGGAGGTCGTCGGTCGGTCT......................................................................................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| ..................................................................................................................................................CGTCGGCCGGTCCAGCGGAGGT................................................................................... | 22 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 |
| ......................................................CCCCGTCGTCGGTCGGTCCAGTC.............................................................................................................................................................................. | 23 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droAna3 | scaffold_13150:2209-2459 - | dan_2218 | GGCCAGGGGGTTGCGCCCCCTCCAGCAGCCGGCCAGGTCGACTCCTCCAGGTTAGGGGCAGCAGCCAGCCAGGTCACCTCCTCCAGCAGCCAGCCAGATCGTCCCCTCCAGGTTAGGAACAGAGGCCAGCCAGATCACCTCCTCCAGCAGCCGGCCAGGTCGCCTCCACAAGNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN |
| Species | Read alignment | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
Generated: 05/18/2015 at 01:47 AM