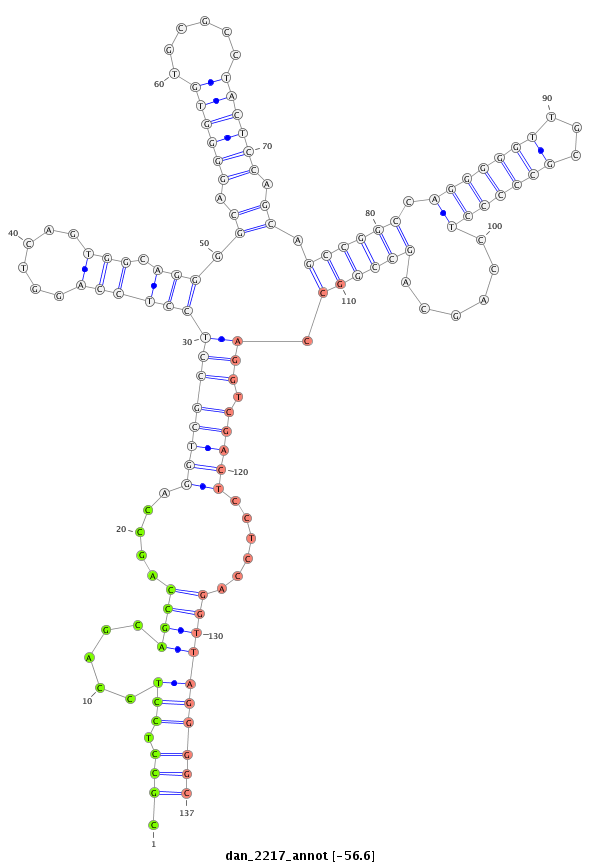

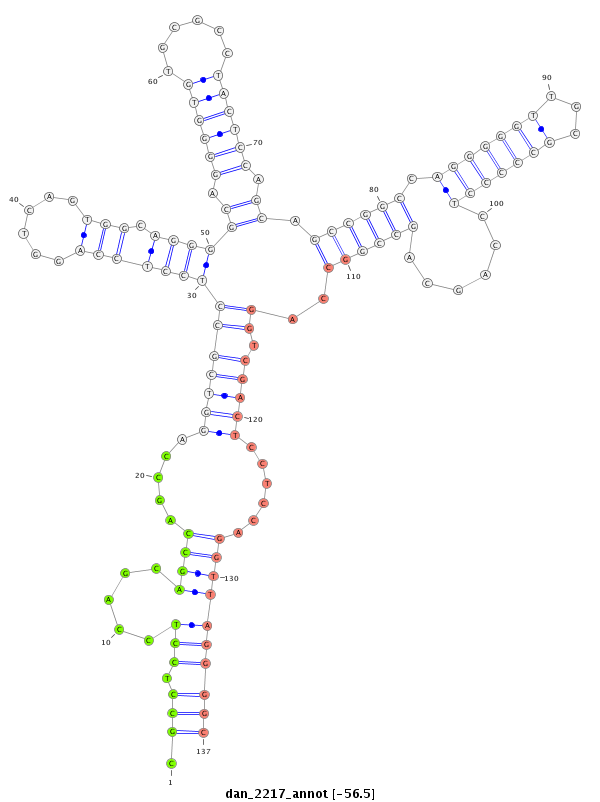
ID:dan_2217 |
Coordinate:scaffold_13150:934-1084 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
No conservation details. |
| -56.5 | -56.5 | -56.2 |
 |
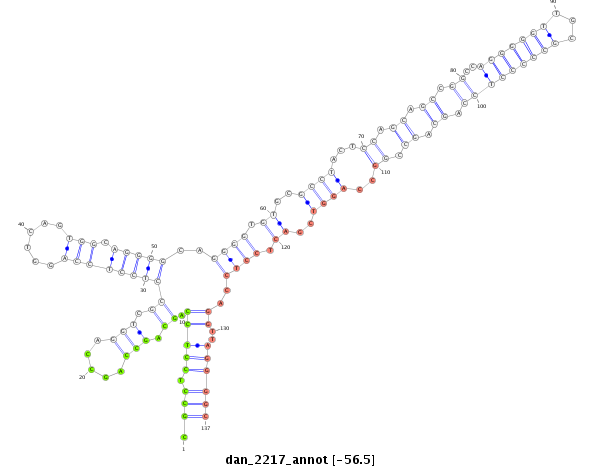 |
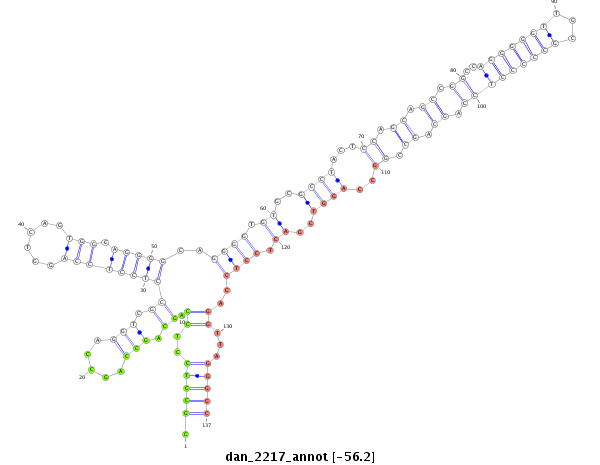 |
exon [dana_GLEANR_20368:11]; CDS [Dana\GF18978-cds]; intron [Dana\GF18978-in]
No Repeatable elements found
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## CCTCCTGCCGGCCAGGGACAGGGGCAGGGGTGTGCGCCTCCTCCAGCAGCCGTCCAGGTCGCCTCCTCCAGCAGCCAGCCAGGTCGCCTCCTCCAGGTCAGTGGCAGGGGCAGGGGTGTGCGCCTACTCCAGCAGCCGGCCAGGGGGTTGCGCCCCCTCCAGCAGCCGGCCAGGTCGACTCCTCCAGGTTAGGGGCATCAGCCAGCCAGGTCACCTCCTCCAGCAGCCAGCCAGATCGTCCCCTCCAGGTT ***********************************************************.(((.(((.....((((.....((((((((((((((......))).))).((.((((((......)))))).)).((((((.(((((((...)))))))......)))))).))).)))))......))))))))))******************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | M058 embryo |
V039 embryo |
V106 head |
M044 female body |
V055 head |
|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................................................ATCAGCCAGCCAGGTCACCTC.................................. | 21 | 0 | 18 | 0.33 | 6 | 6 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................AGGTTAGGGGCATTAGCCAGCC............................................ | 22 | 1 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................CAGCCAGGTCACCTCCTCC.............................. | 19 | 0 | 20 | 0.20 | 4 | 0 | 0 | 4 | 0 | 0 |
| ....................................................................................................................................................................................................ATCTGCCAGCCAGGTCACCTC.................................. | 21 | 1 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................TCAGCCAGCCAGGTCACCTC.................................. | 20 | 0 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................CGCCTCCTCCAGCAGCCAGCC........................................................................................................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| .....................................................................................................................................................................................CTCCAGGTTAGGGGCATT.................................................... | 18 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................ATCAGCCAGCCAGGTCACC.................................... | 19 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................GCCAGGTCGACTCCTCCAGGTTAGGGGC....................................................... | 28 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ....................................................................................................................................................................................................ATAAGCCAGCCAGGTCACCTC.................................. | 21 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
|
GGAGGACGGCCGGTCCCTGTCCCCGTCCCCACACGCGGAGGAGGTCGTCGGCAGGTCCAGCGGAGGAGGTCGTCGGTCGGTCCAGCGGAGGAGGTCCAGTCACCGTCCCCGTCCCCACACGCGGATGAGGTCGTCGGCCGGTCCCCCAACGCGGGGGAGGTCGTCGGCCGGTCCAGCTGAGGAGGTCCAATCCCCGTAGTCGGTCGGTCCAGTGGAGGAGGTCGTCGGTCGGTCTAGCAGGGGAGGTCCAA
*******************************************************.(((.(((.....((((.....((((((((((((((......))).))).((.((((((......)))))).)).((((((.(((((((...)))))))......)))))).))).)))))......))))))))))*********************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M058 embryo |
V106 head |
M044 female body |
V105 male body |
|---|---|---|---|---|---|---|---|---|---|
| ........................................................CCAGCGGAGGAGGTCGTC................................................................................................................................................................................. | 18 | 0 | 20 | 0.40 | 8 | 8 | 0 | 0 | 0 |
| .................................................................GAGGTCGTCGGTCGGTCCAG...................................................................................................................................................................... | 20 | 0 | 20 | 0.20 | 4 | 0 | 0 | 4 | 0 |
| ....................................................AGGTCCAGCGGAGGAGGTCGTCG................................................................................................................................................................................ | 23 | 0 | 20 | 0.10 | 2 | 1 | 1 | 0 | 0 |
| ....................................................................................................................................................................................................................TGGAGGAGGTCGTCGGTCGGT.................. | 21 | 0 | 20 | 0.10 | 2 | 0 | 2 | 0 | 0 |
| .................................................................................................................................................................CGTCGGCCGGTCCAGCTG........................................................................ | 18 | 0 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................AGTCGGTCGGTCCAGTGGAGG................................. | 21 | 0 | 18 | 0.06 | 1 | 0 | 1 | 0 | 0 |
| .....................................................................................................................................................................GGCCGGTCCAGCTGAGGAGG.................................................................. | 20 | 0 | 19 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| ......................................................................................................................................................................................................................GAGGAGGTCGTCGGTCGGTCT................ | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ............................CCACACGCGGAGGAGGTCGTC.......................................................................................................................................................................................................... | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ....................................GGAGGAGGTGGTCGGCAGGT................................................................................................................................................................................................... | 20 | 1 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| .........................................................................................................................CGGATGAGGTCGTCGGCCC............................................................................................................... | 19 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ...................................................CAGGTCCAGCGGAGGAGGTC.................................................................................................................................................................................... | 20 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| ...................................CGGAGGAGGTCGTCGGCAGCG................................................................................................................................................................................................... | 21 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 |
| .................................................................................................................................................................................TGAGGAGGTCCAATCCCCGT...................................................... | 20 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
| ...................................................................................................................................................................................................GTAGTCGGTCGGTCCAGTGG.................................... | 20 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| .................................................................................CCAGCGGAGGAGGTCCAGTCAC.................................................................................................................................................... | 22 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................AGTCGGTCGGTCCAGAGGAGG................................. | 21 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| .....................................................................................CGGAGGAGGTCCAGTCACCG.................................................................................................................................................. | 20 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droAna3 | scaffold_13150:884-1134 - | dan_2217 | CCTCCTGCCGGCCAGGGACAGGGGCAGGGGTGTGCGCCTCCTCCAGCAGCCGTCCAGGTCGCCTCCTCCAGCAGCCAGCCAGGTCGCCTCCTCCAGGTCAGTGGCAGGGGCAGGGGTGTGCGCCTACTCCAGCAGCCGGCCAGGGGGTTGCGCCCCCTCCAGCAGCCGGCCAGGTCGACTCCTCCAGGTTAGGGGCATCAGCCAGCCAGGTCACCTCCTCCAGCAGCCAGCCAGATCGTCCCCTCCAGGTT |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
Generated: 05/18/2015 at 01:47 AM