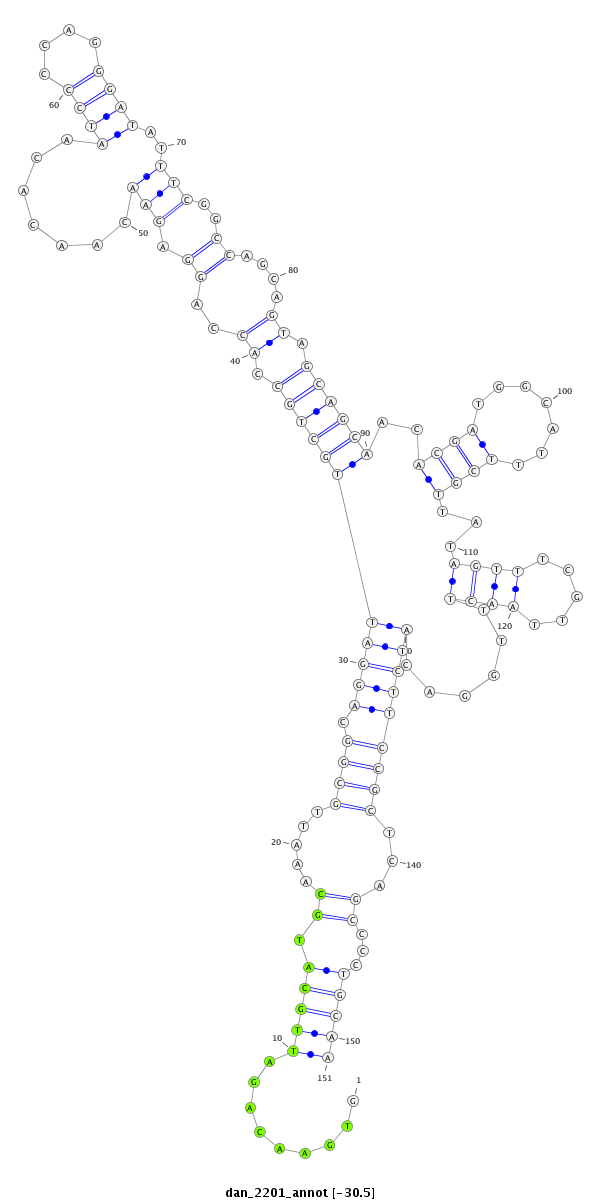
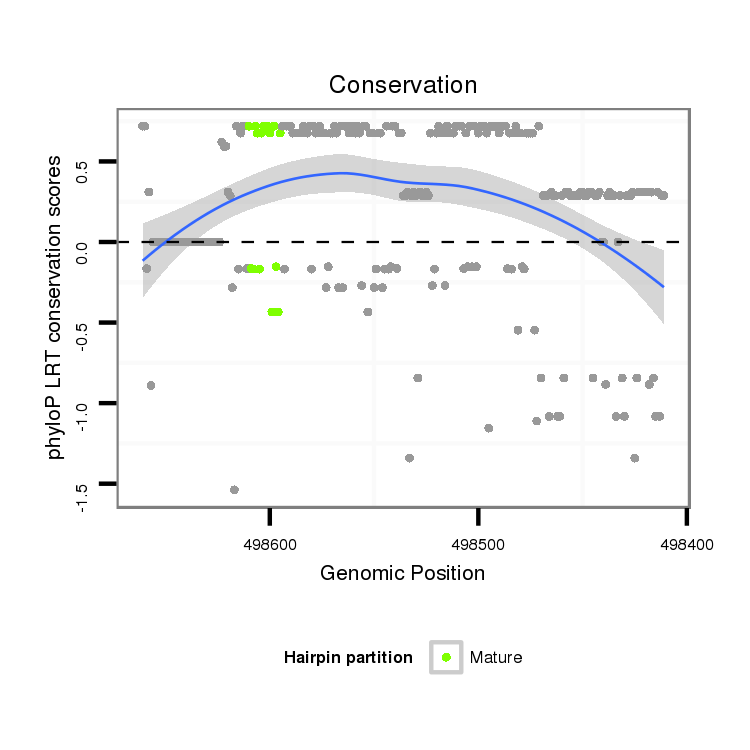
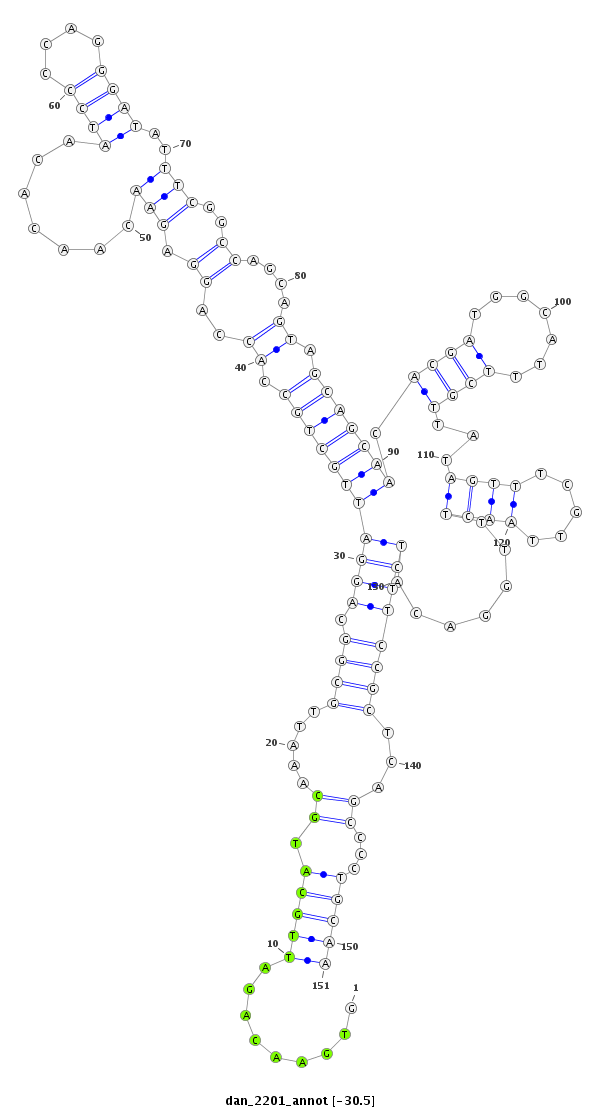
ID:dan_2201 |
Coordinate:scaffold_13335:498461-498611 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -30.5 | -30.0 | -30.0 |
 |
 |
 |
exon [dana_GLEANR_21486:1]; CDS [Dana\GF19445-cds]; intron [Dana\GF19445-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- ATGATGGTGGAATGGTGGACCAGGGAGGGCAGTCGACGTGGAGGGAACATGTGAACAGATTGCATGCAAATTGCGGCAGGATTGCTGCCACCAGGAGAACAACACAATCCCCAGGGATATTTCGGCCAGCAGTAGCAGCAACACGATGGCATTTCGTTATAGTTTCGTTAACTTTGGACATCTTCCGCTCAGCCCCTGCAACGAACGGCCTTGCAATCCAGGACTCCATCTACCCAGGAAAAAAATAAACG **************************************************.........(((((.((.....((((.(((((((((((.((..((.(((.......((((....))))..)))..))....)).))))))..((((.......))))...((((.....))))......)))))))))...))...)))))************************************************** |
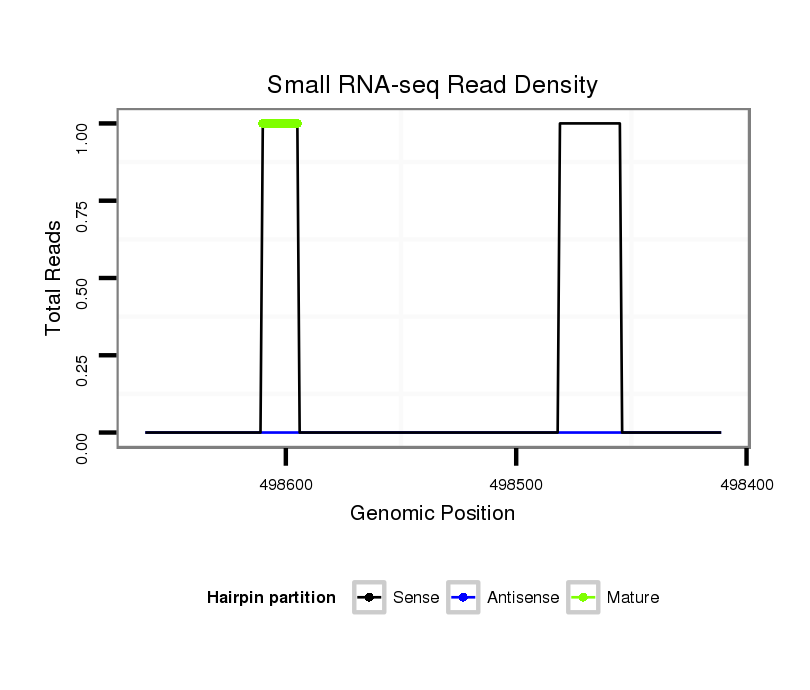
Read size | # Mismatch | Hit Count | Total Norm | Total | M044 female body |
V055 head |
V039 embryo |
|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................................TCTTCCGCTCAGCCCCTGCAACGAACG............................................ | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 |
| ...................................................TGAACAGATTGCATGC........................................................................................................................................................................................ | 16 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 |
| .............................................................................................................................................................TATAGTTTAGGTAACTTT............................................................................ | 18 | 2 | 20 | 0.15 | 3 | 3 | 0 | 0 |
| ...........................................................................................................................................................................................CTCAGCACCTGCAACGACCT............................................ | 20 | 3 | 9 | 0.11 | 1 | 1 | 0 | 0 |
| ..GAGGGTCGAATGGTGGAC....................................................................................................................................................................................................................................... | 18 | 2 | 9 | 0.11 | 1 | 0 | 1 | 0 |
| .....................................................................................................ACACAATCCCCAGGCATT.................................................................................................................................... | 18 | 2 | 12 | 0.08 | 1 | 0 | 0 | 1 |
| .......................................................................................CAACCAGGAGAACACCAC.................................................................................................................................................. | 18 | 2 | 16 | 0.06 | 1 | 0 | 0 | 1 |
| ...................................................................................GTTGCCATCAGGAGAACTA..................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 |
| ......................................GAGAGGGAACAAGTGAACA.................................................................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 |
|
TACTACCACCTTACCACCTGGTCCCTCCCGTCAGCTGCACCTCCCTTGTACACTTGTCTAACGTACGTTTAACGCCGTCCTAACGACGGTGGTCCTCTTGTTGTGTTAGGGGTCCCTATAAAGCCGGTCGTCATCGTCGTTGTGCTACCGTAAAGCAATATCAAAGCAATTGAAACCTGTAGAAGGCGAGTCGGGGACGTTGCTTGCCGGAACGTTAGGTCCTGAGGTAGATGGGTCCTTTTTTTATTTGC
**************************************************.........(((((.((.....((((.(((((((((((.((..((.(((.......((((....))))..)))..))....)).))))))..((((.......))))...((((.....))))......)))))))))...))...)))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M044 female body |
M058 embryo |
V105 male body |
V055 head |
|---|---|---|---|---|---|---|---|---|---|
| ............ACCACCCGGTCCCTCCTG............................................................................................................................................................................................................................. | 18 | 2 | 20 | 0.65 | 13 | 13 | 0 | 0 | 0 |
| .................................................................................................................................................................................TGTAGAAGGCGCGTCCGG........................................................ | 18 | 2 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 |
| ....................................................................................................................................................................................AGAAGGCGAGTAAGGGGCGT................................................... | 20 | 3 | 10 | 0.10 | 1 | 0 | 0 | 1 | 0 |
| ......................................................................................................GTGTTGGGGGTCGGTATAAA................................................................................................................................. | 20 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 1 |
| ............................................CAAGTACACTTGTCAAACGT........................................................................................................................................................................................... | 20 | 3 | 15 | 0.07 | 1 | 0 | 1 | 0 | 0 |
| ....................................................................................................................................................................................AGAAGGCGAGTAAGGGGCG.................................................... | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droAna3 | scaffold_13335:498411-498661 - | dan_2201 | ATGATGGTGGAATGGTGGACCAGGGAGGGCAGTCGACGTGG-------------------------------------------------------------AGGGAACATGTG-AACAGATTGCATGCAAATTGCGGCAGGATTGC----TGCCACCAGGAGAACAACACAATCCCCAGGGATAT--------TTCGGCCAGCAGTA------------GCAGCAACACGATGGCATTTC-------GTTATAGTTTCGTTAACTTTGGACATCTTCCGCTCAGCCCCTGCAACG-AACGGCCTTGCAATCCAGGACTCCATCTACCCAGGAAAAAAATAAACG |
| droBip1 | scf7180000396494:2340-2619 + | ATGAA------------------------------------------ATTGGAGAACGGGGCGAAGGACAGCTGGGGGGAGGCCATAGGTGGCAAGACACCAAGGCAACATGTG-AACAGATTGCATGCAAATTGCGGCAGGATTGC----TGCCACCAGGAGAACAACACAATCCCCAGGGATAT--------TTCGGCCACCAGCAGCAGCAGCAACAGCAGCAACACGATGGCATTTC-------GTTATAGTTTCCTTAACTTTGGACACCTTCCGCCTAACCCATGCCCCAAAACGGCCTTGCAACCCA--TCTCCC-CCCCCCACAAAAAATACCACCG | |
| droRho1 | scf7180000777155:9504-9674 - | ATA-----------------------------------TGGAGAGAAATCG-----------------------------------------------------TTATCATATAATACTGATTGGACCCATATTGCGGCAGGAATGCAGACTGCAGCCAGTATAACAACACCATGCCATGGTTTTCAGCTAGGTTTTGG-------------------------CCTCACGCTGGCATTTTGCTCTGCGCTGTGGTTTCGTTAACTTTAGTCACCCACCGCCAA------------------------------------------------------------- |
| Species | Read alignment | |||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
|||||||||||||||||||||||||||||||||
| droBip1 |
|
|||||||||||||||||||||||||||||||||
| droRho1 |
|
Generated: 05/18/2015 at 04:01 AM