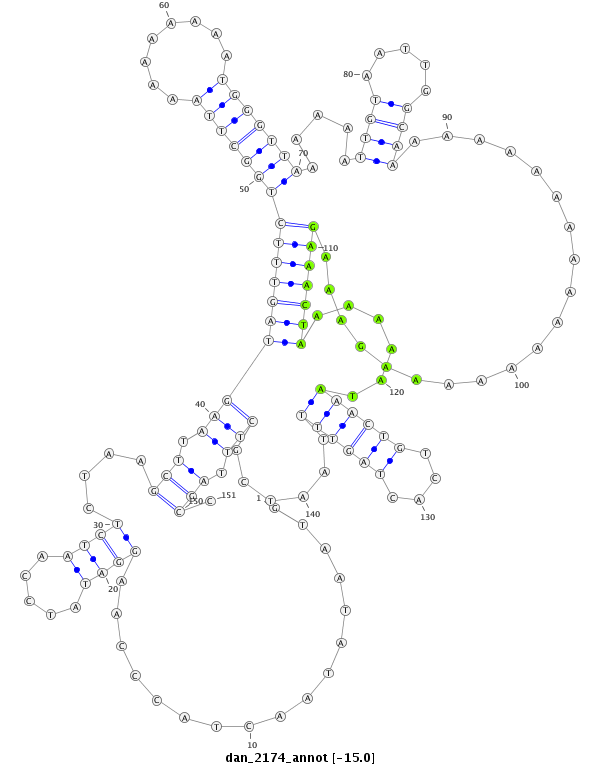
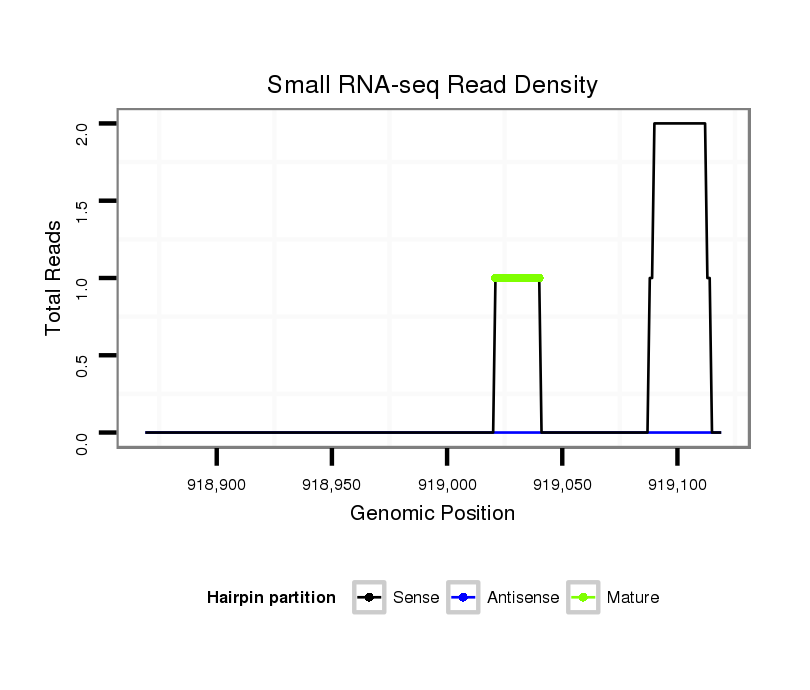
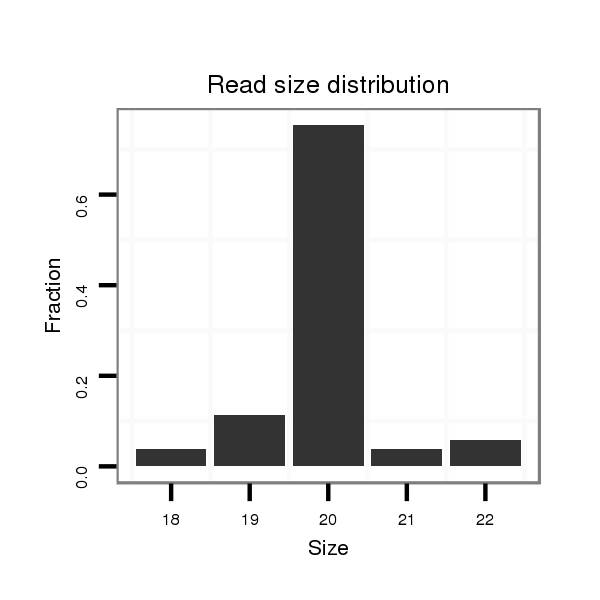
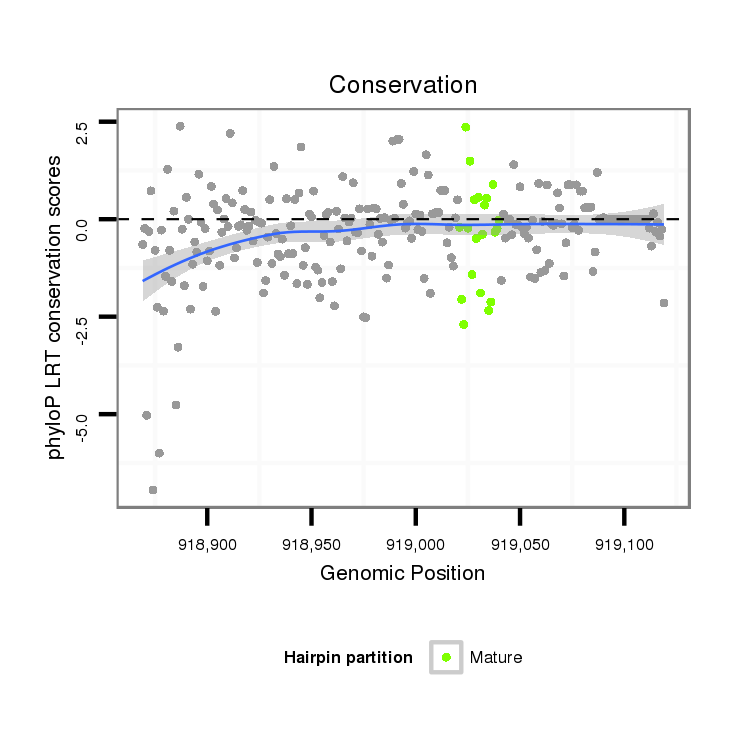
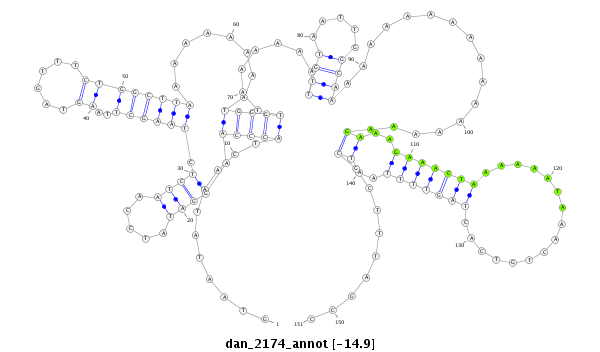
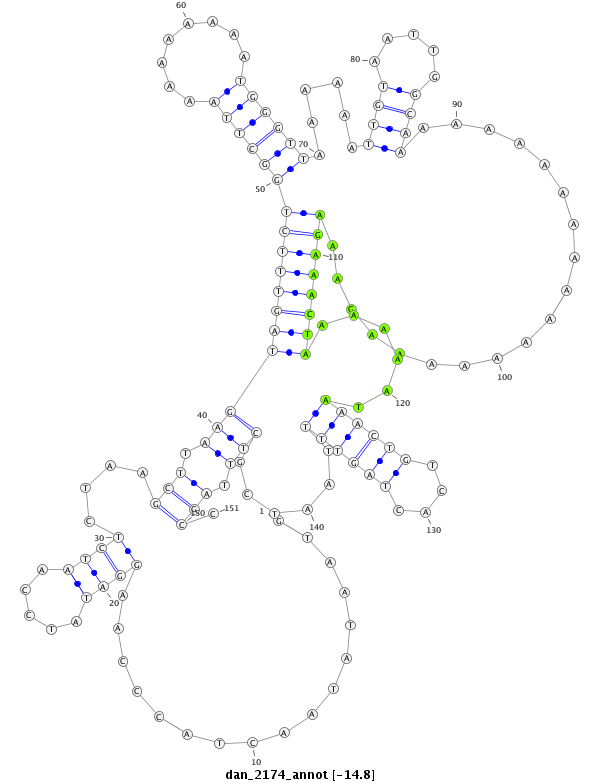
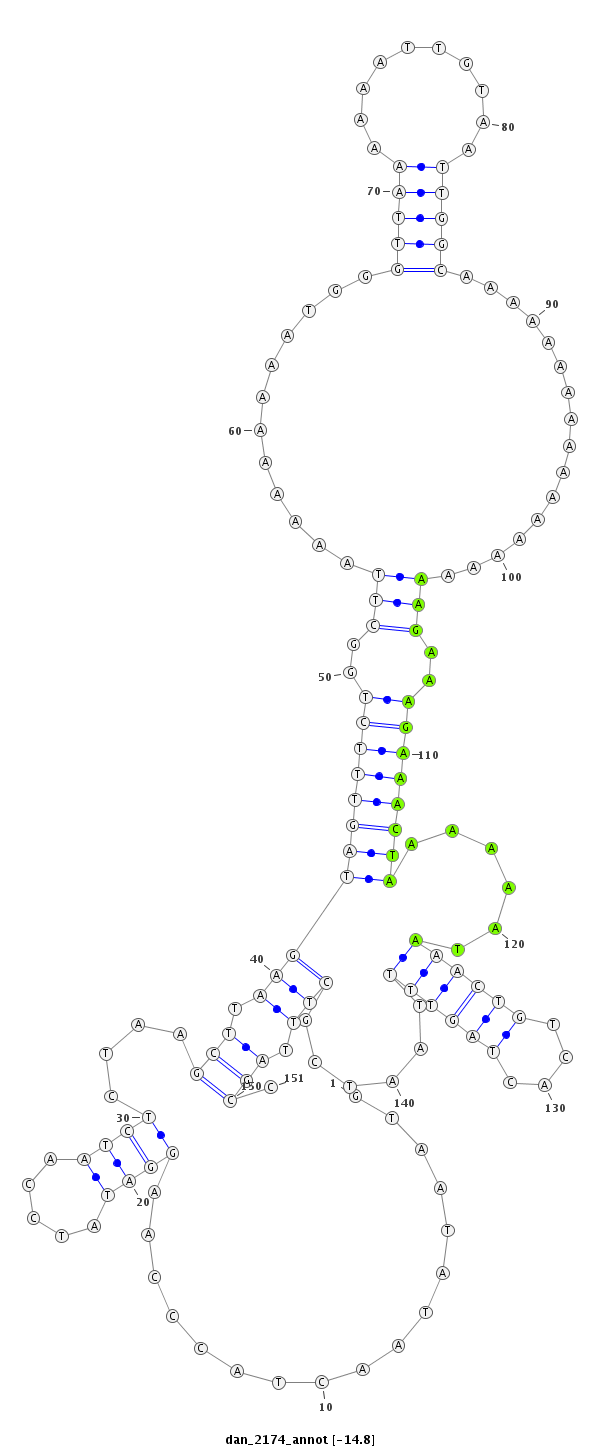
ID:dan_2174 |
Coordinate:scaffold_13334:918919-919069 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -14.9 | -14.8 | -14.8 |
 |
 |
 |
CDS [Dana\GF21834-cds]; exon [dana_GLEANR_5732:1]; intron [Dana\GF21834-in]
| Name | Class | Family | Strand |
| (A)n | Simple_repeat | Simple_repeat | + |
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- CACCACCTCCTCCAACTCCTCCTCCAACAACTCCACCAACAACAATAGCGGTAATATAACTACCCAAGGATATCCAATCTCTAAGCTTAAGTAGTTTCTGGCTTAAAAAAAAATGGGTTAAAAAATTGTAATTGGCAAAAAAAAAAAAAAAAAAGAAAGAAACTAAAAAATAAACTGTCACTAGTTTTAATCGCTTTAGCCTTGATTTAGTTGTCAGGCAGGAAGGCAGGTGTCCTTTGGGATTCCTGTCA **************************************************.................((((.....))))....(((.(((((((((((((((((........))))))).....((((.....))))....................)))))))......((((((....))))))......))).))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M058 embryo |
V105 male body |
V055 head |
V039 embryo |
V106 head |
|---|---|---|---|---|---|---|---|---|---|---|
| ........................................................................................................................................................AAGAAAGAAACTAAAAAATA............................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................AAAAAAAAAAAAAAAAGAAAGACACTAAAAAA................................................................................. | 32 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| .............................................................................................................................................................................................................................GAAGGCAGGTGTCCTTTGGGATT....... | 23 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................AGGAAGGCAGGTGTCCTTTGGGATTCC..... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................AAAAAAACAAAAAAAAAAGAAAGAAACC....................................................................................... | 28 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................AAAAAACAAAAGAAAGAAACT....................................................................................... | 21 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................AAAAGACAGAAACTAAAAAA................................................................................. | 20 | 1 | 3 | 0.33 | 1 | 0 | 0 | 0 | 1 | 0 |
| ............................................................................................................................................GAAAAAAGAAAAAAGAAAGAAACT....................................................................................... | 24 | 2 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................AAAAAATACAAAAGAAAGAAACT....................................................................................... | 23 | 2 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................TGTAATTGGCAGAGTAAAAAA........................................................................................................ | 21 | 3 | 13 | 0.15 | 2 | 0 | 2 | 0 | 0 | 0 |
| ................................................................................................................................................AAAAAAAAAAGAAAGAAACC....................................................................................... | 20 | 1 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................GAAAAAAGAACGAAACTAA..................................................................................... | 19 | 2 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................AAGGCAGGTGTCCTAATGG.......... | 19 | 3 | 10 | 0.10 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...............................................................................................................................................AAAAAAAAAAAGAAAGAAA......................................................................................... | 19 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................AAAAAAAAAAAAAAAGAAACTAAAAA.................................................................................. | 26 | 1 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................AAAAAGAAAAAAAAAAAGAAAGA........................................................................................... | 23 | 1 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................AAAAAAAAAAAAAAAGAAAGAA.......................................................................................... | 22 | 0 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................ATAAAAAAGAACGAAACTAAGAA.................................................................................. | 23 | 3 | 14 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................AAAAAAAAAAAAAAAAAGAAAGAAAG........................................................................................ | 26 | 1 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................AATTAGCAAAAAAAAAGAAAAAAA.................................................................................................. | 24 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................AAAAAAAAAAAAAAAAGGAAGAACC........................................................................................ | 25 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................AAAAAAAACAAAAAGAAAGA........................................................................................... | 20 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................AAAAAAGAAAAAAAAAGAAAGA........................................................................................... | 22 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................AAAAAAAAAACAAGAAAGAAACGA...................................................................................... | 24 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................AAAAAAAAGATAAAAAAAGAAAGAA.......................................................................................... | 25 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................AAAAAAAAGAAAGAAACC....................................................................................... | 18 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................CATTTTCCAACAACTCCACCA..................................................................................................................................................................................................................... | 21 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................AAAAAAAAAAAAGAAAGAA.......................................................................................... | 19 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................AAAAAAGAAAAAAGAAGGAAAC........................................................................................ | 22 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................ATTGGCAAAAGTAAAAAGAAAA................................................................................................... | 22 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| ................................................................................................................................................AAAAAAAAAAGAAAGAAA......................................................................................... | 18 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................AAAAAAAAAAAAAAAAGAAAG............................................................................................ | 21 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................AAAAAAAGAAAGAAACCA...................................................................................... | 18 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................AAAAAAAAAGAAAGAAACC....................................................................................... | 19 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................AATTGGCAACAACAAAAAAAAAAAA................................................................................................. | 25 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
|
GTGGTGGAGGAGGTTGAGGAGGAGGTTGTTGAGGTGGTTGTTGTTATCGCCATTATATTGATGGGTTCCTATAGGTTAGAGATTCGAATTCATCAAAGACCGAATTTTTTTTTACCCAATTTTTTAACATTAACCGTTTTTTTTTTTTTTTTTTCTTTCTTTGATTTTTTATTTGACAGTGATCAAAATTAGCGAAATCGGAACTAAATCAACAGTCCGTCCTTCCGTCCACAGGAAACCCTAAGGACAGT
**************************************************.................((((.....))))....(((.(((((((((((((((((........))))))).....((((.....))))....................)))))))......((((((....))))))......))).))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M058 embryo |
V055 head |
M044 female body |
V039 embryo |
V106 head |
|---|---|---|---|---|---|---|---|---|---|---|
| ........GGAGGTTGAGGAGGAGGTT................................................................................................................................................................................................................................ | 19 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ................................................................GTTACTATAGGTTAGACATT....................................................................................................................................................................... | 20 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................CGATAGGTTAGAGGTTCG..................................................................................................................................................................... | 18 | 2 | 7 | 0.43 | 3 | 2 | 0 | 1 | 0 | 0 |
| .......AGGAGGTTGAGGAGGAGG.................................................................................................................................................................................................................................. | 18 | 0 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 | 0 |
| .........................TTGTTGAGGTGGTTGATT................................................................................................................................................................................................................ | 18 | 2 | 17 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................GGTGGTTGGTGTTTTCGACA....................................................................................................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| ........................................................................................................................................TTTTTTTTTTTTTTTTTTCT............................................................................................... | 20 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................TTTTGAGGCGGTTGTGGTTA............................................................................................................................................................................................................. | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ................ATGAGGACTTTGTTGAGGT........................................................................................................................................................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ......GATGAGGTTGACGAGGAGG.................................................................................................................................................................................................................................. | 19 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droAna3 | scaffold_13334:918869-919119 + | dan_2174 | CA--CCACC-TCCTCCA-ACT-------------------------------------------------------------------------------------------CCTCCTCCAACAA------------------------------------------C---TCCACCA-----ACA---ACAATAGCGGTAAT-------------ATA--------------ACTAC-------------------------------------------------------------------------------------CCA----AGG-----------------------------------------------------------ATATCCAATCTCTAAGCTTAAGTA---------GTTTCTGGCTTA----------------------------A----------------------------------------------------------------------------------------------------------AA-AAAAATGG---------GT-TAA-------AAAATTGTAATTG-----G----------------------------CAA-----------AAAA------------------AAAAAAAAAA-------------------------------AAGA---AA---------------------------G-A----AAC-TAAAAAA---------------TAA------------------ACTGTCACTAGTTTTAATCG-------------CTTTAGCCTTGATTTAGTTGTCAGGCAGGAAGGCAGGTGTCCTTTGGGATTCCTGTC---------------A |
| droBip1 | scf7180000396733:560434-560651 + | CA--ACAAT-TCAAACA-ATC-------------------------------------------------------------------------------------------ACACCACCAACAA------------------------------------------T---AATAGCG--------------------GTAAT------------------------------G---C-------------------------------------------------------------------------------------CCC----AGG-----------------------------------------------------------ATAACTAAATCCTAAAAAAAACAAAATACTATTATTTGCGGCTAAAAAGGGTATTTTTTA------ATATTAGTATTGT-----------------------------------------------------------------------------------------AATTG----------------------------------------------------------GCAACAACAACAAAAAAAA------------------------------------------AAAT----TGTA---------------------------------------------------------------------------AA-----TT-------------------GT------------------T------------ATTTTAGAATAAGACGAAATATATAGCCTGGATTTAGTTGTCACAC--G---------------------TAGTGCC---------------T | |
| droKik1 | scf7180000302441:609301-609523 - | CA--CCGTT-AACGGCA-ACA-------------------------------------------------------------------------------------------ACAATTCGCACGT------------------------------------------C---GACGCTC-----ACAACGGCAGAAGCAGCAGC------------------------------GCCAG-------------------------------------------------------------------------------------CCGAAGACAC-----------------------------------------------------------ATAACGGAGCTCTGTA----------------------------------------------------------------------------------------AAATGGGCGTTAAGAAGCATATCAGCGAAACAC----------------------------------------CATGTGAGTAAATA---------CA-GCA-------TAAATAAAAAACA-------------------------A----------ACTAAAAACATACTTAAA-------------AA-----AAAAAA-------------------------------AAGA---A---------------------------ACAAACAAA-----AACAG----------------------------------------ACGCTATTTTTGA-------------------------------------------------------------------------------TTAACTTGGC | |
| droFic1 | scf7180000453807:1030449-1030615 + | ACTAGC----AACAACA-CAG-------------------------------------------------------------------------------------------GCAGTAGGA------------------GAATTTATAATAAGTAACTCTGCGGCTGC------------------------------------------------------------------------------------------------------------------------------------------------------TGCT----------------------------------------------------------------------------------------------------------------TCG----------------------------A----------------------------------------------------------------------------------------------------------TC-GAGAGTTG---------CG-CAAAAAAAAGCAAA--------AAAAAAAGA----------------------------A------------------------------AAA----AAGC--GAG------------------------------GA---G---------------------------AAAAACAAA-----AGCAACTGCCGCAAATCTTCCAC--------TTAATTATAAA----------------------------------------CTTATTTA----------------------------------------------------TTTGG | |
| droEle1 | scf7180000490564:229229-229396 + | AG--ACAAC-AACCGCA-ATC-------------------------------------------------------------------------------------------CCGCCACACACAC------------------------------------------A---AACGCCC-----GAA---ACACTCGCAGACAT-------------CCG--------------CACAC-------------------------------------------------------------------------------------CCA----CA-----------------------------------------------------------------------------CACACACAAGTTCACTACGTCACGCACA--------------------------------------------------------------------------------------------AAC--------ACAAACCATGTCTGCCAATA--------------------------AGTTTGC------------------------------ACA--------------------------AAA-----------AAAA--------------AAAA----TGAAAA-------------------------------AGGA---A---------------------------AC-A----AAC-AAAAAAA--------------------------------------------------------TAGA---------------------------------------------------------------------------------- | |
| droRho1 | scf7180000779318:129229-129437 - | CA--ACATT-ACATACA-ACA-------------------------------------------------------------------------------------------GCAACAATAGCATACAAACCAGCAACAACAGCAGTAATATAGC-----------------------------------CAGAAGCAGCAGC------------------------------AAC-------------------------------------------A-------CCAAGAACAACA--------------ATTGCAGCA-------------------------------------------------------------------------------------------------------ATATCACC--------------------------------------------------------------------------AGAAAACACACCAGC------------------AA--------------------------------------------------------------------------------------------------------------CAA-----------CATCACCA------------------ATAATG-------------------------------TACA---AAAA-------AAAACAAAAAAAAACA---------A-----------------------GCTAC------------------A----CATTGGCGACAATCG-------------CTTTTGC-TCAATGCATTT-------------------------------------------------GTCAG | |
| droBia1 | scf7180000302075:2368769-2368915 + | AC--CGATT-CCCAGCT-TCA------------------------------------------------------------------------------------------GACTTCAGTGGC------------------------------------------------------C----AACA---GCCAG--------------CTAAATATCCA--------------C-----------------------------------------------------------------------------------------------GTTC----------------GTGC-----------------------------------------GT--------------------------------------------------CGGTTTTTTGTGGCCAAATCGGTGC-----------------------------------------------------------------------------------------GGC----------------------------------A----------GAAAAT--------------------------AAATATCACACACAA-----------CAAAAAAA------------------AAAGAA-------------------------------AAGA---AA---------------------------T-A----GAA-TCATAAC--------------------------------------------------------AAAA---------------------------------------------------------------------------------- | |
| droTak1 | scf7180000410451:13995-14160 + | GC--CCAAC-TCA-CCG-CCC---ACGCCAC--------------------------------------------------------------------------------------ACGAGCAG---------------------------------------------C------A-----GGA---GC-------------------------------------------------------------------------------------------------------------------------------------AGAAGGTGA-----------------------------------------------------------GTGTCTAAATCTTATAA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------TT---------CCAAAATGATTTTAAAATAAAA-----------------------------------------------------------------------AAA----TGTTGA-------------------------------AGAA---T---------------------------TC-A----TTC-TAAATAT-------------TATAAAACAATTATAAAATCTAAA----C--------------AATA--------TTTAAAA-------------------C--T---------------------CTG----------CTCGTTTTAA | |
| droEug1 | scf7180000409095:477919-478120 + | CT--TCAAC-TTCAACT-GCA-------------------------------------------------------------------------------------------GCAGCAACAACAG------------------------------------------T---GACAAAA------GG---GCAATTA--------------------------------------------------------------------------------------------------------------------------------------AGT-----------------------------------------------------------AGGCCCAAGATCTAGGCAAAAAGA------------------------------------------------------------------GTTAAGAGCCTCTAAGATGAGCGACGGGGTTAAGGCAATCAGATTTG----------------------------------------CTTGT--------------------------------AAA--------TTGAAAAGA----------------------------A------------------------------TTA----AAGT--TAA------------------------------TA---A---------------------------GAAAAGATA-----A--------------------------ACTTTAAAGTTTAAA---------------------------------------------------------C--T---------------------TTGAATATTGAAATCTAGTTAGC | |
| dm3 | chrX:9164765-9164939 - | CA--GGAGC-AACAGCA-ACT------------------------------------------------------------GCAATTGAAGCCACATCATCTGCAGCAACT-TCAACTTCAACTG------------------------------------------T---T------------------------------------------------------------------------------------------------------------------------------------------------GCAGCAGCAGCAAAAGA-----------------------------------------------------------AGGCCCAAATACCACAGAAAACTCAAATTATATACC------------------------------------------------------------------------------------------------------------------------------------------------------------------TGC------------------------------AGA--------------------------AAA---------------------------------A------------AACATAGATTTGTAGTCCCCTAATCATAG---CAAG--------------------------AAC-----TA-ACA-ATAAA--------------------------------------------------------CTGA---------------------------------------------------------------------------------- | |
| droSim2 | 3r:15722743-15722954 + | CA--CCACC-ACCA-CTAC-CACCACAA--CTACAACAACAACAACAACCAGTACAAGCACCAACAACAACATCAACAACAACAACAAGAACCTCATCGACGGCGTCAATT-GCGAC------------------------------------------------------------------------------------------------------------------------------------------------------------------G-------ATGGCTGCCATAAATACGGTAAGTACGCGACATCATC----------------------------------------------------------------------------------------------------------------A----------------------------A----------------------------------------------------------------------------------------------------------CC-AAATAATG---------CT-AAAATATAATAAAA--------TAAAAGGAA----------------------------A------------------------------GAA----AAGC--AAA-----------------------------GAAAAG-------------------------------A----A-----AATAA--------------------------------------------------------AAGA---------------------------------------------------------------------------------- | |
| droSec2 | scaffold_68:56829-56988 - | CA--TCATC-TGCAGCA-ACT-------------------------------------------------------------------------------------------TCAACTGCAGCTG------------------------------------------T---TGCAGAA-----ACA---GCAGTTGCACCAGC-------------------------------------------------------------------------------------------------------------------------AGCAGAAGA-----------------------------------------------------------ATGCCCAAATCCCACAGAAAACTGAAATTATCTACC-------------------------------------------------------------------------------------------------------------------------------------------TGCAGAA---------AA-------------------------------------------------------------------------------------------------------A------------GCCATAGATTTGTAGTCCTCTAATCATAG---CAAA--------------------------AAC-----TA-ACA-ATAAA--------------------------------------------------------CTAA---------------------------------------------------------------------------------- | |
| droYak3 | X:12592124-12592305 - | CA--GCAAC-TACAGCA-CCA-CCGTGGCGT--------------------------------------------------------------------------------------ATGCGTAA---------------------------------------------T------AAGCGAACA---ACAAT--------------GTGAACATGAG--------------C-------------------------------------------------------------------------------------------------------------------------------------------------------------------------GTAATATCATCAA----------------------------------------------------------------------------------------------GTTACGACCATCCAATTTCAGGGCTAGATCATAAAATATATCTAGC-----------CTTTAAA---------AAAATTTTGG------------------------------ATA--------------------------AAA--------------------------------A----CATC--AAA-----------------------------CTAAAGA----------AACTCA------------------GA-----AT-------------------AA------------------G------------CCTTTAAAATA---------------------------------------------------------------------------------- | |
| droEre2 | scaffold_4770:931491-931671 - | CT--GATTT-CCCC-TTGC-CATTACCC--TTTGAAAGCCAAA----------------------------------------------------------------------------------------------------------------------------------CACAC-----ACT-----------------------------------------------------------------------------------------------------------------------------------------------------GCGCTCAAAATAATATCTTTTACATTACATTAAATAATGAAAAAGAACCAGTTAACTA-----------------------------------------------------------------ACTCAAGTAGTGTAGCTGCAAGAAAATAACAAGTCCCAAAA---------------------------------------------------------------------------------------AGATTG----------------------------------------------------------------------------------------------------AAAAAA-------------------------------TTCA---AA--AAAATAA----------------AG--C----T-----GTTAA--------------------------------------------------------CTGT---------------------------------------------------------------------------------- | |
| dp5 | XR_group6:8981994-8982186 + | CA--GCAGC-AACAACA-ACA-------------------------------------------------------------------------------------------ACAACAACAACAC------------------------------------------C---ATCACCA-----TCA---GCAATCATCGCAGC------------------------------CCC-----------------------------------------------------------CA-----------------------------C---AAAATGTGCTCAGCATAGTGAGT-----------------------------------------GT--------------------------------------------------TTTTTTTTTTTGGCAAAGGCAAAGC-----------------------------------------------------------------------------------------AGCCACCAACAAAA---------AA---------AAAATTATAATAAAATCAACAACA-----G----------------------------CAG-----------AAAA------------------A------------AATATAAAA--------------AA---A---AAGA---------------------------AA-A----CAC-TAAAAAA--------------------------------------------------------TAGA---------------------------------------------------------------------------------- | |
| droPer2 | scaffold_24:700971-701150 - | AA--GCAACAACACACC-ACA------------------------------------------------------------------------------------------GACAACAACCGC------------------------------------------------------A--------------------ATCCCGCCACACTAACACACACACACACACCCAAACAT-------------------------------------------G-------GCAATGGCTACG--------------GCTACGGCT-------------------------------------------------------------------------------------------------------ACGTCACGCACA--------------------------------------------------------------------------------------------AACACAAAAACAAAAACAATGTCTGCCAATA--------------------------AGTTTGC------------------------------AAA--------------------------AAA-----------GGAA---------------AAA----AAAA---------------------------------A---AAGG--------------------------AAA-----AA-A----T-------------------GA------------------AA---CG-------------TTAC---------------------------------------------------------------------------------- | |
| droWil2 | scf2_1100000004909:9548218-9548381 + | AA--CAACC-TACTACT-ACT-------------------------------------------------------------------------------------------ACTACTACTACTA------------------------------------------C---TACAACA-----ACA---ACAACAACA---AT-------------CCA----CATAGTTTA-ACTGG-------------------------------------------------------------------------------------CGA----TGG-----------------------------------------------------------AGTTCAAACTTCAAAGAG------------------------------------------------GCAATTACTG-----------------------------------------------------------------------------------------------------------AGTAATGA---------CT-GCA-------AAAATCCTAAATA------------------------------------A-----------CAAA---------------TAACAAACAGA------------------------------------A---A---------------------------TT-A----TAT-AAAAACC--------------------------------------------------------AAAA---------------------------------------------------------------------------------- | |
| droVir3 | scaffold_12970:6439346-6439541 + | CA--ACAAC-TTCAACA-GCA-------------------------------------------------------------------------------------------GCAACAACAACAACTTCAGCAGCAGCAGCAGCAACAACAACAACAGCTTCAGCTGC-----------------------------------------------------------------TCT-------------------------------------------A-------TCAGCAGCAGAA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------GATGC-----------------------------------------------------------------------------------------AGC----------------------------------A----------------------------GCAATGACAAAAGAAGAAACCGCAAACAA-----------CAACAACG------------------AAAAAA-------------------------------AAGA---AACA-------AACACACAAGAAAGTT---------A-----------------------TCAAC--------TAATTGATAAA---------------------------------------------------------C--G---------------------CTGCATA-----------CTTAA | |
| droMoj3 | scaffold_6680:6345233-6345436 - | CA--GCAAT-TGCAGCA-GCC-------------------------------------------------------------------------------------------ACAGCAACAGCAA------------------------------------------C---AACAGCA-----GCA---GCAGAAGCAGCAAC------------------------------ATCATTATCAACTTCATAATGAGCATTGATTGAAGCTAACTGTTGCACTTCTGCTTGGCTGCTG-----------------------------C--------------------------------------------------------------------------------------------------------------AAAAAGTGTTTGAAA------TTAAAAGCTGTGC-----------------------------------------------------------------------------------------AGCCAACAA----------------------------------------------------------------------------------------------------AAACAAAAAAAGGGAAAAA----TAAA--GA-----------------------------G---AAGA---------------------------AA-AATAGA-----AAAAA--------------------------------------------------------CTGT---------------------------------------------------------------------------------- | |
| droGri2 | scaffold_14906:8318006-8318189 + | CA--CAAT--AATGACA-ACA-------------------------------------------------------------------------------------------GCATCAACAACGG------------------------------------------TGGCAGCAACA-----ACA---GCGGAAACAACAAC------------------------------AACAATGCCAA--------------------------------------------------------------------------------------------------------------TAAAGATGCAATCGATGG-----------------------CAGAAACTCAAATTG------------------------------------------------GTAAGAGCCGTGC-----------------------------------------------------------------------------------------AGCTTATGGTATTT--------------------------------AAA--------AATAAAAGA----------------------------A------------------------------AAA----AGAC--AAAAAAAA-----------------------AAAA---GA--AAAGA-------------------AA-T----TA--------------------------------------------------CG-----------------------------------------------------------------------------------------TTAGCTTTAA |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/18/2015 at 03:57 AM