
ID:dan_2132 |
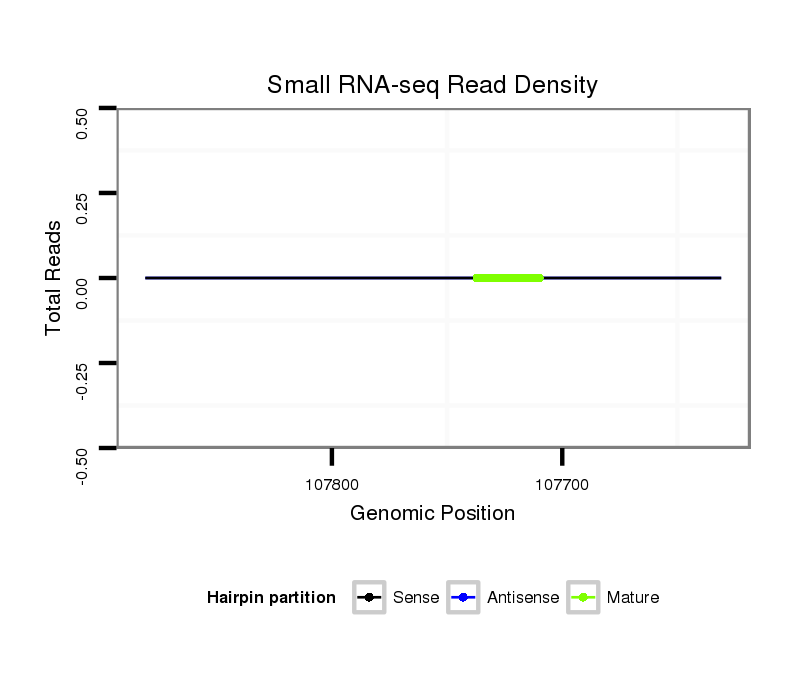
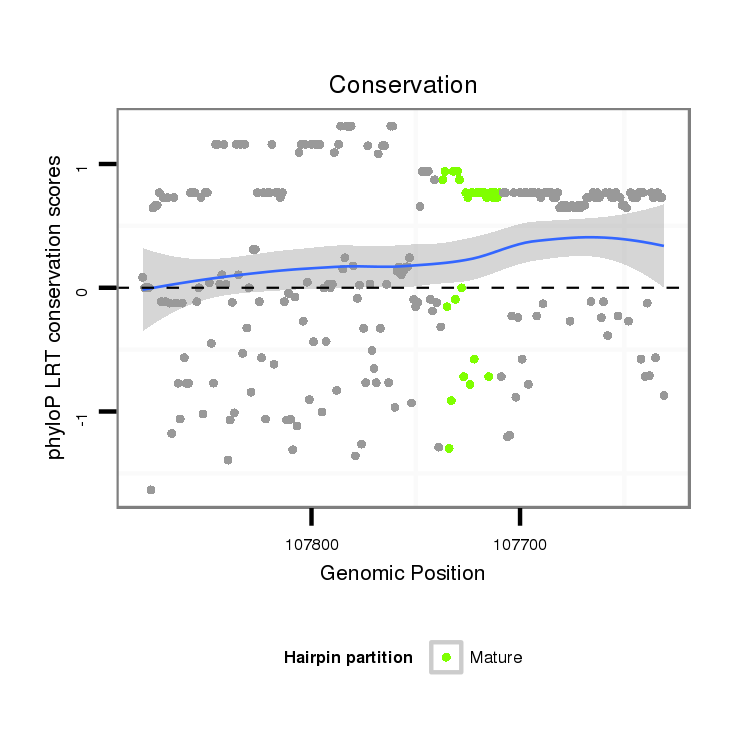
Coordinate:scaffold_13272:107681-107831 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -20.9 | -20.6 | -20.5 | -20.5 |
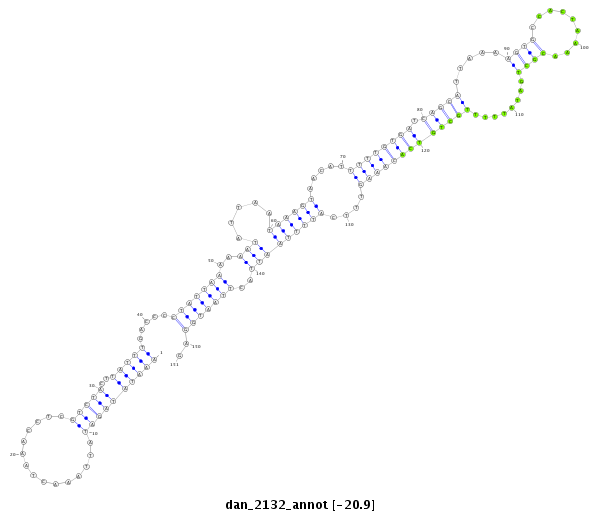 |
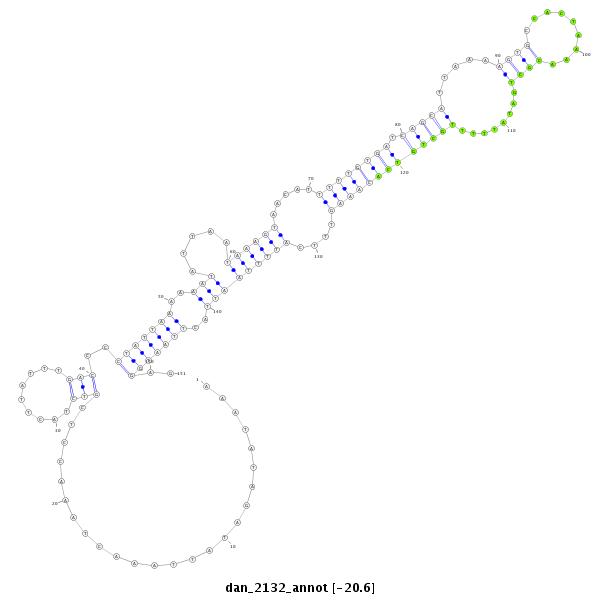 |
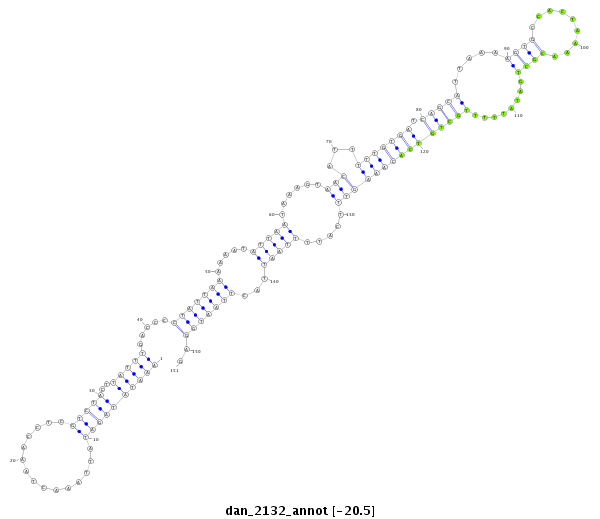 |
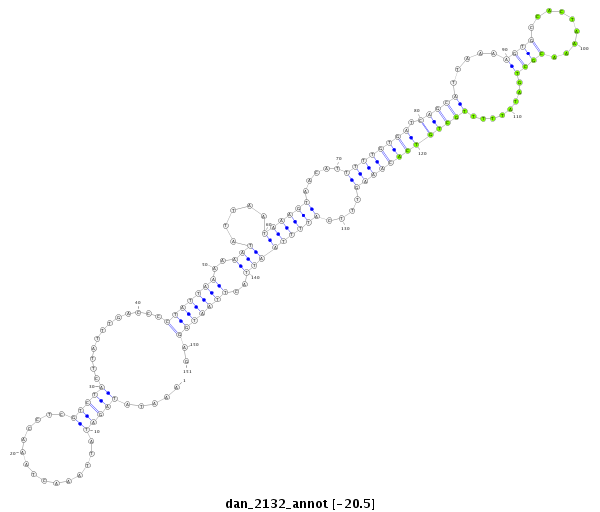 |
exon [dana_GLEANR_3486:2]; CDS [Dana\GF20609-cds]; intron [Dana\GF20609-in]
No Repeatable elements found
| mature | star |
| ---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## AAATGCATTTGAGCGGCTACGTTTTTAGGGTTAGATTCAACCACAAAATAAAATATAGATATTAAACTAAACCTCGTCTACTTATTTGACCCCTATTAAAAAATATTAATAAAGTAACATTTTTGTGATCAGCATTAAAAGTGCCACTAAAACGCTGATATTTTTGCTGTCACAAAGTTTCATTTTAATTACTTAATGGAGTCTGAAGCTGAAGCTACTGGATCTGCTTCAAGTAGGGAAAGGATGCTGCG **************************************************................((........................((((((......(((((......(((...(((((((.(((((.....((((.........))))........))))))))))))))).....)))))....))))))))************************************************** |
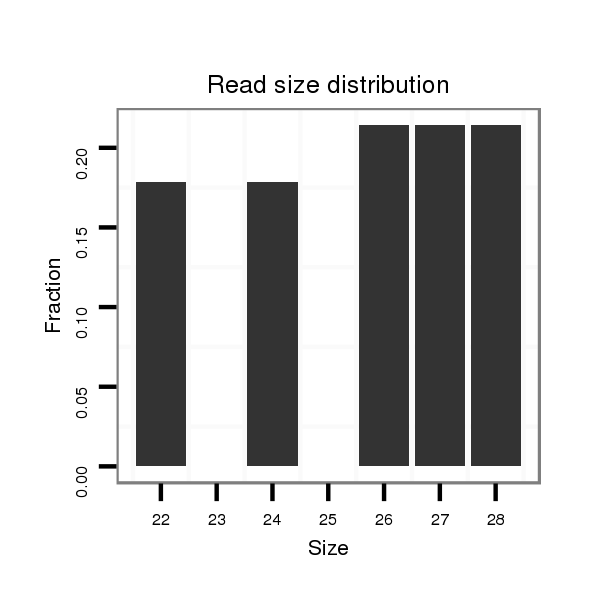
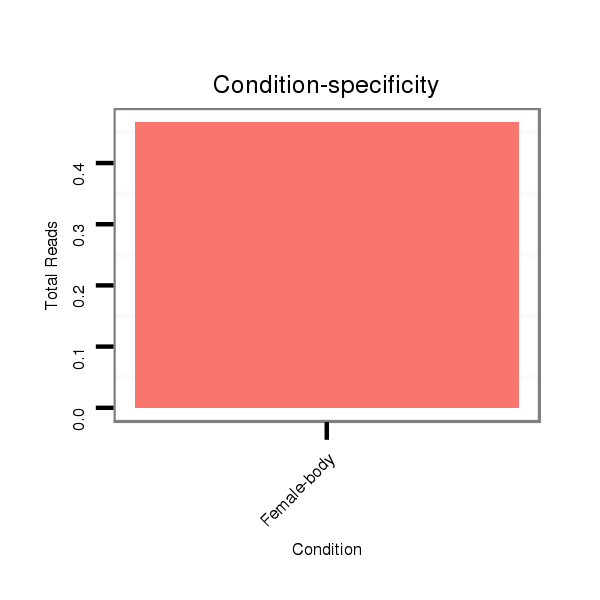
Read size | # Mismatch | Hit Count | Total Norm | Total | M044 female body |
M058 embryo |
V055 head |
V039 embryo |
V105 male body |
V106 head |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................AACCTTTTTGTGATCACCA..................................................................................................................... | 19 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................................................................TAATGGAGTCTGAAGCTGAAGCTACT................................ | 26 | 0 | 5 | 0.40 | 2 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................TCTGAAGCTGAAGCTACTGGATCTGCTT...................... | 28 | 0 | 9 | 0.33 | 3 | 2 | 0 | 0 | 0 | 1 | 0 |
| .............................................................................................................................................................................................TACTTAATGGAGTCTGAAGCTGAAGCTAC................................. | 29 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................AACCTTTTTGTGATCAACA..................................................................................................................... | 19 | 2 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................................................................................................................................AATTACTTAATGGAGTCTGAAGCTGAAG..................................... | 28 | 0 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................TGGAGTTTGAAGCAGCAGCTAC................................. | 22 | 3 | 6 | 0.17 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................................TAATTACTTAATGGAGTCTGAAGC.......................................... | 24 | 0 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................AACGCTGATACTTATACTGT................................................................................. | 20 | 3 | 15 | 0.13 | 2 | 1 | 0 | 0 | 0 | 0 | 1 |
| .....................................................................................................................CATTTTAGTGAGCAGCAT.................................................................................................................... | 18 | 2 | 8 | 0.13 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................TCTGAAGCTGAAGCTACTGGATCTGC........................ | 26 | 0 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................CACTAAAACGCTGATATTTTTGCTGTCA............................................................................... | 28 | 0 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................CACTAAAACGCTGATATTTTTGCTGT................................................................................. | 26 | 0 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................TAAAACGCTGATATTTTTGCTGTCACA............................................................................. | 27 | 0 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................GCGTCTACTTATTTGACCCCTATTAAAA...................................................................................................................................................... | 28 | 1 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........TGAGCGGCTACGTTTTTAGGGTTAG......................................................................................................................................................................................................................... | 25 | 0 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................TACTGGATCTGCTTCAAGTAGGGAA........... | 25 | 0 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................ATCTGAAGCTACTGGATCTGCTTCAAG.................. | 27 | 1 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................TGGATCTGCTTCAAGTAGGGAAAGGA....... | 26 | 0 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................................TGGATCTGCTTCAAGTAGGGAAAGG........ | 25 | 0 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................TACTGGATCTGCTTCAAGTAGGG............. | 23 | 0 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......TGTGAGCGGCTACGTTTTTAGGGC............................................................................................................................................................................................................................ | 24 | 2 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .................TACGTTTTTAGGGTTAGATTCAACCAC............................................................................................................................................................................................................... | 27 | 0 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................CCACTAAAACGCTGATATTTTTGC.................................................................................... | 24 | 0 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................TTTTTAGGATTAGCTGCAACCA................................................................................................................................................................................................................ | 22 | 3 | 12 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................CACTAAAACGCTGATATTTTTG..................................................................................... | 22 | 0 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................TTTAGGGTGAGGTTCAACC................................................................................................................................................................................................................. | 19 | 2 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................................TACTGGATCAGCTTGAAGT................. | 19 | 2 | 16 | 0.06 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...............................TAGATTCATCCGCAAAAAAAAA...................................................................................................................................................................................................... | 22 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................ACTGGATCTGGTTGAAGT................. | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................GATCTGCTTCGAGAAGGG............. | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................TTTTAGGGTGAGGTTAAACC................................................................................................................................................................................................................. | 20 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................AGAGCACCACTAAAACGCT............................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ............................................................................................................................................GTCCCGCGAAAACGCTGAT............................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................ATCTGCTTGAAGGAGGGA............ | 18 | 2 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
|
TTTACGTAAACTCGCCGATGCAAAAATCCCAATCTAAGTTGGTGTTTTATTTTATATCTATAATTTGATTTGGAGCAGATGAATAAACTGGGGATAATTTTTTATAATTATTTCATTGTAAAAACACTAGTCGTAATTTTCACGGTGATTTTGCGACTATAAAAACGACAGTGTTTCAAAGTAAAATTAATGAATTACCTCAGACTTCGACTTCGATGACCTAGACGAAGTTCATCCCTTTCCTACGACGC
**************************************************................((........................((((((......(((((......(((...(((((((.(((((.....((((.........))))........))))))))))))))).....)))))....))))))))************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M058 embryo |
M044 female body |
V106 head |
V055 head |
V039 embryo |
|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................................................................................................................................................CTAGACGAAGTTCATCCCTTTCCTAC..... | 26 | 0 | 11 | 0.27 | 3 | 1 | 2 | 0 | 0 | 0 |
| ....................................................................................................................................................................................GTAAAATTAATGAATTACCTCAGACTT............................................ | 27 | 0 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................GTAATTTTCACGGTGATTTTGCGACT............................................................................................. | 26 | 0 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................CGTAATTTTCACGGTGATTTTGCGACT............................................................................................. | 27 | 0 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................................................................................................................................................CGACTTCGATGACCTAGAGGAAGTTCA................. | 27 | 1 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................................................AAACGACAGTGTTTCAAAGTAAAATTAAT............................................................ | 29 | 0 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................CGACAGTGTTTCAAAGTAAAATTAAT............................................................ | 26 | 0 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................................GACCTAGACGAAGTTCATCCCTTTCCT....... | 27 | 0 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......AAACTCGCCGATGCAAAAATCCCA............................................................................................................................................................................................................................ | 24 | 0 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................CCACGGTGATTTTGCGACTATAA......................................................................................... | 23 | 1 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................ATGCAAAAATCCCAATCTAAGTTGGT................................................................................................................................................................................................................ | 26 | 0 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...............CGATGCAAAAATCCCAATCTAAGTTG.................................................................................................................................................................................................................. | 26 | 0 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............CGCCGATGCAAAAATCCCAATCTAAGT.................................................................................................................................................................................................................... | 27 | 0 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............CGCCGATGCAAAAATCCCAGTCTAAGTT................................................................................................................................................................................................................... | 28 | 1 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 |
| .TTACGTAAACTCGCCGATGCAAAAAT................................................................................................................................................................................................................................ | 26 | 0 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................................................TGACCTAGACGAAGTTCATCC.............. | 21 | 0 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................TAGACGAAGTTCATCCCTTTCTTAC..... | 25 | 1 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................CGCAAAAATCCCAATCTAAGT.................................................................................................................................................................................................................... | 21 | 1 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................TTTCACGGTGATTTTGCGACTATAA......................................................................................... | 25 | 0 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................AGACGAAGTTCATCCCTTTCCTAC..... | 24 | 0 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................CTAGACGAAGTTCATCCCTTTCCT....... | 24 | 0 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................CTAGACGAAGTTCATCCCTTTCCTACG.... | 27 | 0 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................CCTAGACGAAGTTCATCCCTTTCCTAC..... | 27 | 0 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................CCTAGACGAAGTTCATCC.............. | 18 | 0 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................................CTAGACGAAGTTCATCCCTTTCCTA...... | 25 | 0 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................................CCTAGACGAAGTTCATCCCTTTCCTACG.... | 28 | 0 | 11 | 0.09 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................CGGTGATTTTGCGACTATAAAAA...................................................................................... | 23 | 0 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 |
| ................................................................................................................................AGTCGTAATTTTCACGGTGATTTTGC................................................................................................. | 26 | 0 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................TCCCTCACACTTCGACTACG.................................... | 20 | 3 | 13 | 0.08 | 1 | 0 | 0 | 1 | 0 | 0 |
| .......................................................................................................................................................................................................................AGGACCTAGGCCAAGTTCATC............... | 21 | 3 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................TGGTAGCAGACGAATAAACTGGG............................................................................................................................................................... | 23 | 3 | 14 | 0.07 | 1 | 0 | 0 | 0 | 1 | 0 |
| .....GTAAACTCGTCGAGGCAAA................................................................................................................................................................................................................................... | 19 | 2 | 14 | 0.07 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...............................................................................................................................................................................................................CGACTTCGATAACATAAACGAA...................... | 22 | 3 | 17 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................ATTTTGCGACTAGGAAAA...................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| ...........................................................................................................................ACACTAGTCGTAATTTTCACGGTGAT...................................................................................................... | 26 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................ACCATTTTTTATAATTATTT.......................................................................................................................................... | 20 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droAna3 | scaffold_13272:107631-107881 - | dan_2132 | AAATGCATTTGAG-CGGCTACGTT----------TTTAGGGTTA-GATTCAACCACAAAATAAAATA---------------TAGATATTAAACTAAAC-CTCGTCTACTTATT--------TGACCCCTATTAAAAAATATTAATAAAGTAACA------------------------------------TTTTTGTGATCAGCATTAAAAGTGCCACTAAAACGCTGATATTTTTGCTGTCACAAAGTTTCATTTTAATTACTTAATGGAGTCTGAAGCTGAAGCTACTGGATC---TGCTTCAAGTAGGGAAAGGATGCTGCG |
| droBip1 | scf7180000395241:4783-5061 - | A---T---TTGAG-CCGCAGCAAA---------TTTTA-GTTTAAACTTGAACGCCTAAAAA--GTATTGATTCAATACTATTAAACATTGAACTACAACGTTGT-TATTCATT--------CGGCCGCAATTAAAAACTAATCATTGCGTAAGCTATTGATGACCCCAACGTGAGTGCACCCCGTTTTGGTTATTGTGGTTAGT-TTAAAAGTTCCACGGAAAC-TTGTTGTTTTTGTTGTCATAACCTTGCACTTAAATTACTTAATGGA-------------GCTACTGGATCTGATGCTTC----AGGGAGAAGGTGTTGCT | |
| droEug1 | scf7180000408770:6038-6298 + | T---GCATTCGGGGTGGTTTTGTTGCGGAGTTGGTTTG-GTTTTGAATTGAACTCGGAAAAAT-----------AATATTTTTGGAAATTGAACTGCCCCG--GTTTGTTTATT--------ATACAGCAATTAAAATC--G------------------------------------CACCCCATTTGGATTATTGTGATCTGT-TTAAGCGTGCCACGGATAC-TTGTTGTTTTTGTTGTCATAATCGTGAACTTAAATGACATAATGGAGTCTGGAGCTGAAGCCACTGTGTGTGCTGCTGCAAGCAGGGAGAAAATGCTGCG | |
| droEre2 | scaffold_4929:21148637-21148696 + | T---A------------------------------------------------------------------------------------------------------------------------------TCAAAAAAAAAAAAAAAAGAAACA------------------------------------TTTGTCGAATAATTATTAAAAGCAGCAAAAAAAC----------------------------------------------------------------------------------------------------- | |
| droVir3 | scaffold_12958:3154920-3155010 + | A---C--------------------------------------A-AATTGGAACACAAGAAA---------------------------TA-----AAATAAGGT-TAATTATTTCTATTTGTGAACGCTATTGAAAAAACTCAATAGAGCAACA------------------------------------TTTTGGTGACC---------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
Generated: 05/18/2015 at 03:48 AM