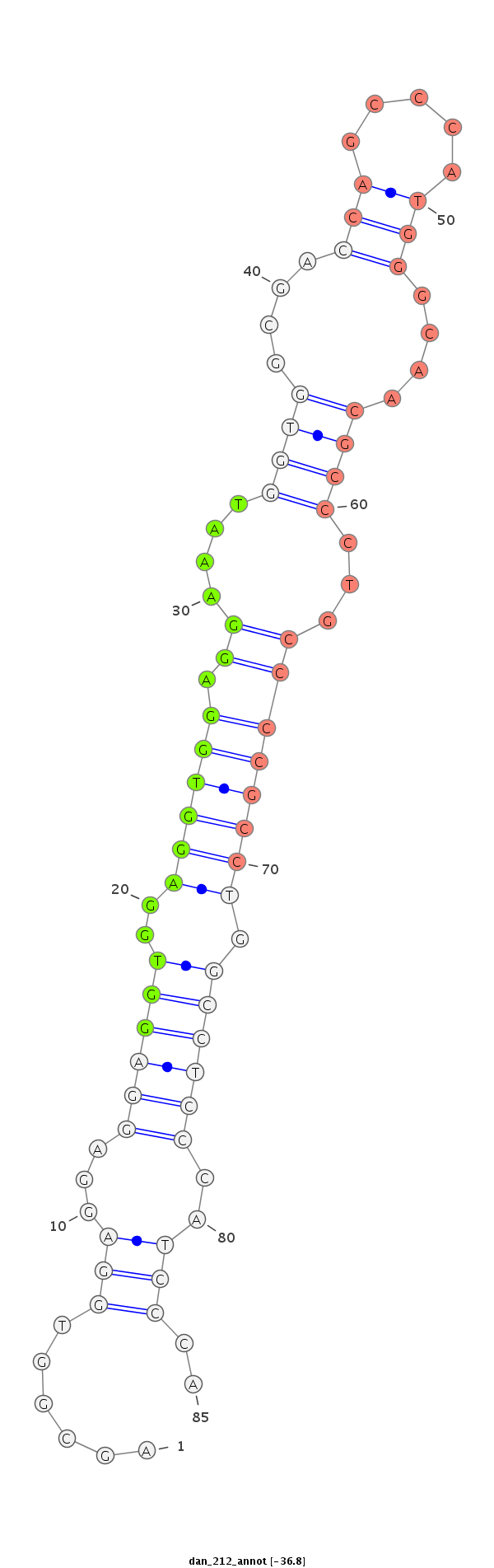
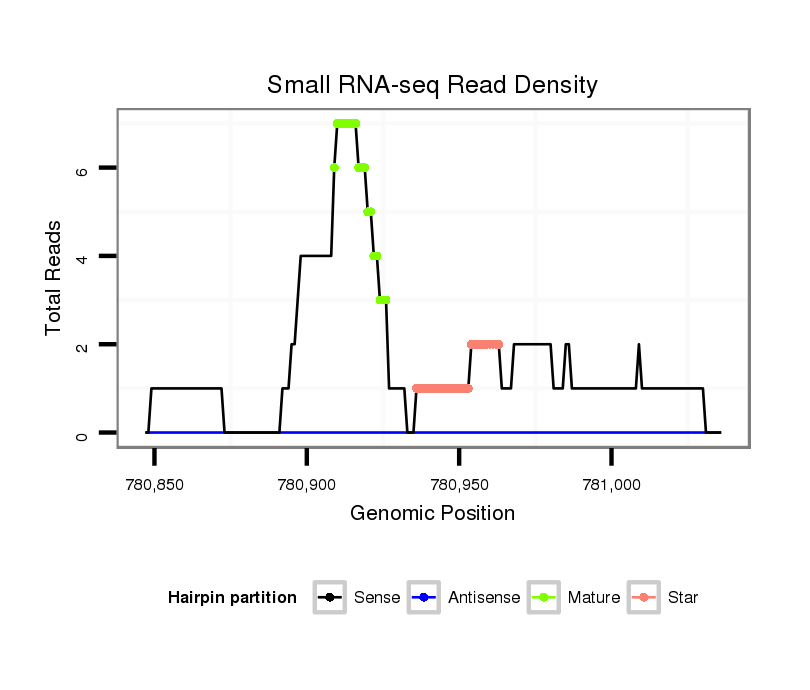
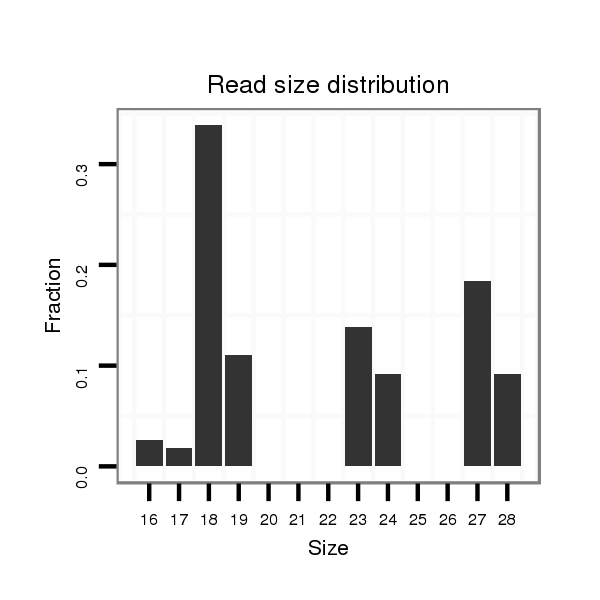
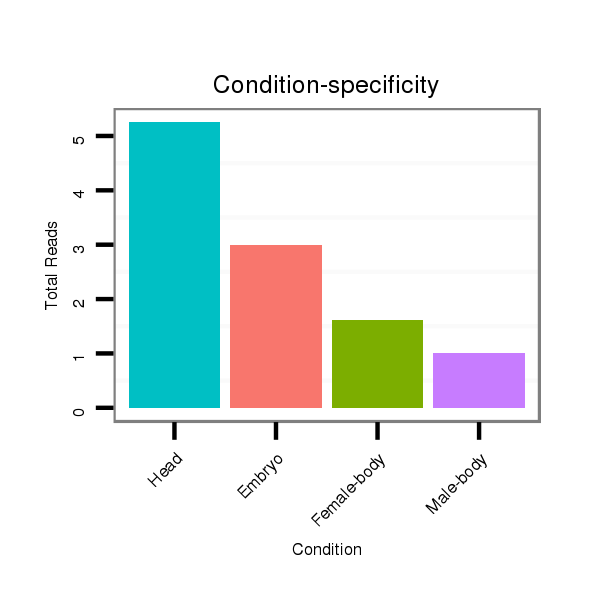
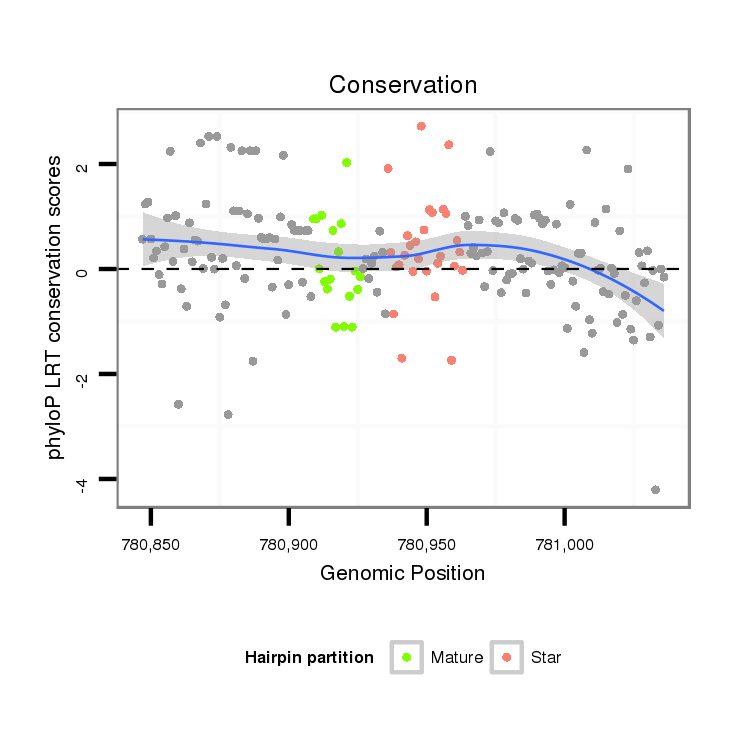
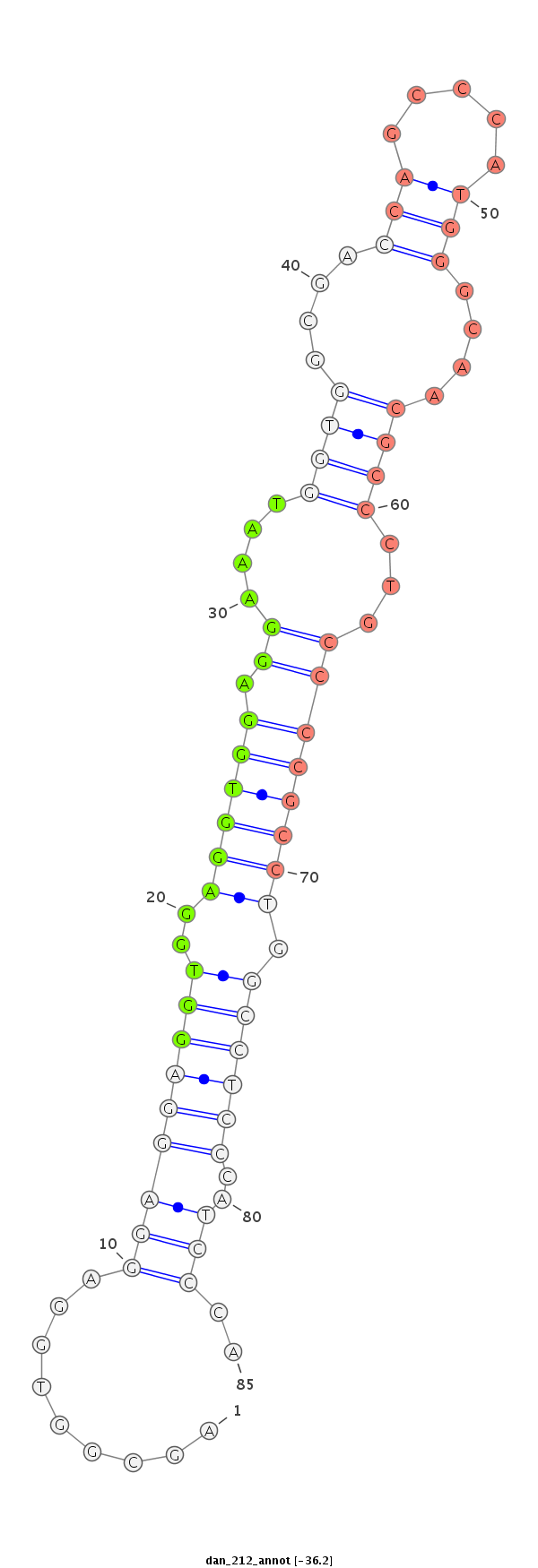
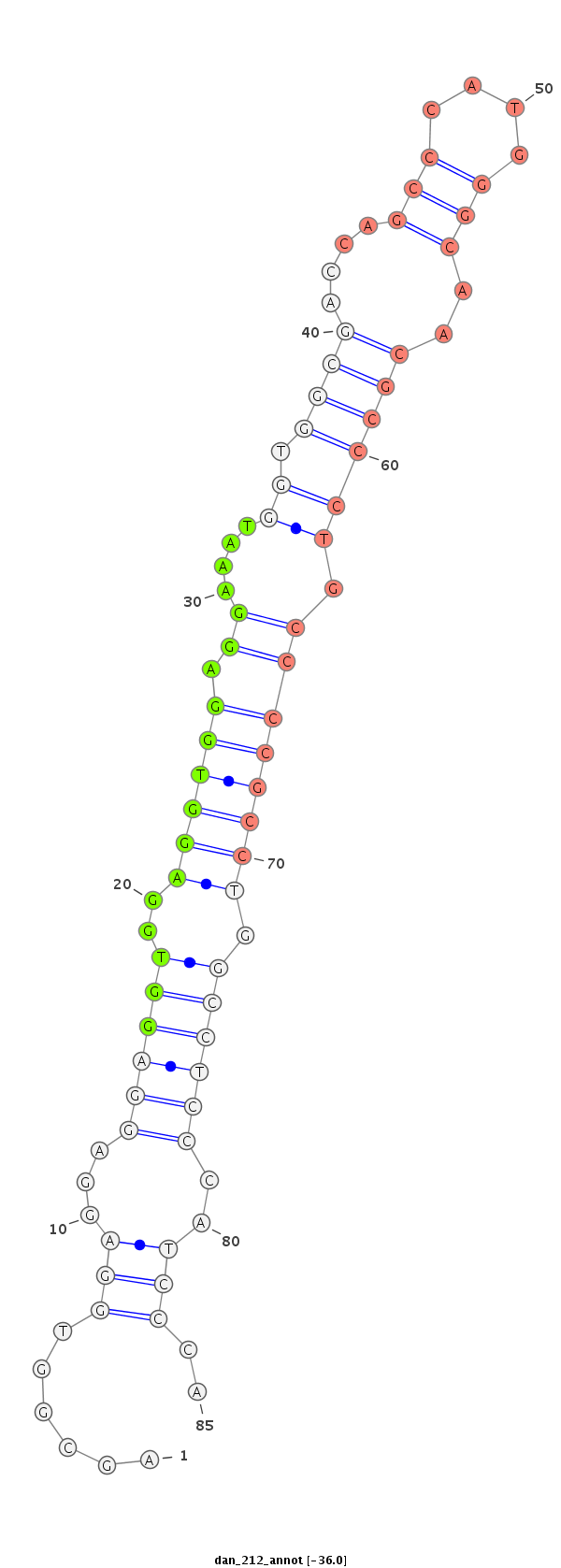
ID:dan_212 |
Coordinate:scaffold_13334:780897-780986 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -36.5 | -36.2 | -36.0 |
 |
 |
 |
exon [dana_GLEANR_5728:8]; CDS [Dana\GF21829-cds]
| Name | Class | Family | Strand |
| (CGG)n | Simple_repeat | Simple_repeat | + |
| ############################################################################################################################################################################################## GCGGCACGCCCACGGCACACAGCGGTGGTGGCGGCGGCGGCGGTGGAAGCGGTGGAGGAGGAGGTGGAGGTGGAGGAAATGGTGGCGACCAGCCCATGGGCAACGCCCTGCCCCGCCTGGCCTCCCATCCCAATGCCAGCAGCAGCAACTCCTCGGTGTCGGGAGGACCGCTTGGCGCCGCTGCCGTAGT ***********************************************......(((...((((((..((((((.((....((((....(((.....)))....))))...)))))))).))))))..)))..********************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V106 head |
M044 female body |
M058 embryo |
V055 head |
V105 male body |
V039 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ..............................................................GGTGGAGGTGGAGGAAAT.............................................................................................................. | 18 | 0 | 1 | 2.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................CAGCCCATGGGCAACGCCCTGCCCCGCC......................................................................... | 28 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................GAGGGGGTGGAGGTGGAGGAAAT.............................................................................................................. | 23 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................GTGGAGGAGGAGGTGGAGGTGGAG................................................................................................................... | 24 | 0 | 2 | 1.00 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGGTGGAGGAGGAGGTGGAGGTGG..................................................................................................................... | 25 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................TGCCAGCATCAGCAACTCCTCGGTGTCG............................. | 28 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................GTGGAGGTGGAGGAAATGGTGGC........................................................................................................ | 23 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..................................................GGTGGAGGAGGAGGTGGAGGTGGAGGA................................................................................................................. | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ...........................................................................................................CTGCCCCGCCTGGCCTCCCATCCCAAT........................................................ | 27 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................GAGGACCGCTTGGCGCCGCTGC...... | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................GCAGCAGCAACTCCTCGGTGTCGGG........................... | 25 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................GAAGCGGTGGAGGAGGAGGTGGAGG........................................................................................................................ | 25 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..GGCACGCCCACGGCACACAGCGGT.................................................................................................................................................................... | 24 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...GCACGCCCACGGCACACAGCGGC.................................................................................................................................................................... | 23 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................CTCCCATCCCAATGCCAGC.................................................. | 19 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................GGCCGCTTGGCGCCGCTGCCGTAGT | 25 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ............................................................GAGGTGGAGGTGGAGGAA................................................................................................................ | 18 | 0 | 5 | 0.60 | 3 | 3 | 0 | 0 | 0 | 0 | 0 |
| .......................GGTGGTGGCGGCGGCGGC..................................................................................................................................................... | 18 | 0 | 11 | 0.55 | 6 | 6 | 0 | 0 | 0 | 0 | 0 |
| .............................................................AGGTGGAGGTGGAGGAAA............................................................................................................... | 18 | 0 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .............................................................................AATGGAGGTGACGAGCCCATGGGC......................................................................................... | 24 | 3 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ...................................................GTGGAGGAGGAGGTGGAGGTTGAG................................................................................................................... | 24 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................GCGGTGGAGGGGGAGGTGGAGGTGGA.................................................................................................................... | 26 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................CGGTGGAGTAGGAGGTGGAGGTG...................................................................................................................... | 23 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................GTGGTGGCGGCGGCGGCGGT.................................................................................................................................................. | 20 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GGTGGAGGAGGAGGTGGAGGTGG..................................................................................................................... | 23 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...........................................................GGAGGTGGAGGTGGAGGA................................................................................................................. | 18 | 0 | 17 | 0.35 | 6 | 0 | 6 | 0 | 0 | 0 | 0 |
| ................................................GCGGTGGAGGCGGAGGTTGAGGTGGAG................................................................................................................... | 27 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................GCGGCGGCGGCGGTGGAAG............................................................................................................................................. | 19 | 0 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................GTGGAGGCGGAGGTGGAGGTGGAGGA................................................................................................................. | 26 | 1 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................GTGGAGGAGGAGGTGGAGGCGGAGGAC................................................................................................................ | 27 | 2 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................GGTGGAGGTGGAGGAA................................................................................................................ | 16 | 0 | 7 | 0.29 | 2 | 0 | 0 | 0 | 2 | 0 | 0 |
| .............................................................AGGCGGAGGTAGAGGAAAT.............................................................................................................. | 19 | 2 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ......................................AGCGGTGGAAGCGGTGGA...................................................................................................................................... | 18 | 1 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..................................................GGTGGAGGAGGAGGTGGAG......................................................................................................................... | 19 | 0 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .............................................................AGGTGGAGGTGGAGGAA................................................................................................................ | 17 | 0 | 5 | 0.20 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ....................................................TGTAGGAGGAGGTGGAGGTGGAGGC................................................................................................................. | 25 | 2 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GGTGGAGGAGGAGGTGGA.......................................................................................................................... | 18 | 0 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................GGTGGCGGCGGCGGCGGT.................................................................................................................................................. | 18 | 0 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......GTGCACGACACACAGCGG..................................................................................................................................................................... | 18 | 3 | 20 | 0.10 | 2 | 0 | 0 | 0 | 2 | 0 | 0 |
| ..................................................GGTGGAGGAGGAGGTGGTGGTGGA.................................................................................................................... | 24 | 1 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ...................................................GTGGAGGAGGAGGTGGTGGTGGA.................................................................................................................... | 23 | 1 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................GGTGGAGGAGGAGGTGTA.......................................................................................................................... | 18 | 1 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................ACACCGGTGGTGAAGGCGGC........................................................................................................................................................ | 20 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..................................................GGTGGAGGGGGAGGTGGAGGTGGA.................................................................................................................... | 24 | 1 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................TGGCGGCGGCGGCGGTGG................................................................................................................................................ | 18 | 0 | 15 | 0.07 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................GGAGGAGGTGGAGGTGGA.................................................................................................................... | 18 | 0 | 16 | 0.06 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................AGGGGGATGAAATTGTGGCG....................................................................................................... | 20 | 3 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ............................................................GAGGTGGAGGCGGAGGAA................................................................................................................ | 18 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................GTCGGAATCGGGAGGACC..................... | 18 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..............................................................................................................................ATCCCGGTGCCAGCAGCCGC............................................ | 20 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .............................................................AGGTGGTGGTGGAGGAA................................................................................................................ | 17 | 1 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
|
CGCCGTGCGGGTGCCGTGTGTCGCCACCACCGCCGCCGCCGCCACCTTCGCCACCTCCTCCTCCACCTCCACCTCCTTTACCACCGCTGGTCGGGTACCCGTTGCGGGACGGGGCGGACCGGAGGGTAGGGTTACGGTCGTCGTCGTTGAGGAGCCACAGCCCTCCTGGCGAACCGCGGCGACGGCATCA
**********************************************************......(((...((((((..((((((.((....((((....(((.....)))....))))...)))))))).))))))..)))..*********************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V055 head |
V105 male body |
M044 female body |
M058 embryo |
V106 head |
|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................................................AGGTACCCGTGGCGGGACG............................................................................... | 19 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................GTGGCGGGACGGGGCGGACTG..................................................................... | 21 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................TGTGTCGCCACCATCGTCCCC......................................................................................................................................................... | 21 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................TCGCCACCACCGCCGCCG........................................................................................................................................................ | 18 | 0 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..............................................................................................................................................................AGCCATCCTTGCGAACCG.............. | 18 | 2 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................AGCCATACTGGCGAACCG.............. | 18 | 2 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................TCCATCACCGCCGCCGCCGCCACCT............................................................................................................................................... | 25 | 2 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 | 0 |
| ........................................................CGTCCTCCAACTCCACCTTCTT................................................................................................................ | 22 | 3 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................AGGGTAGGGTTGCGGTTG.................................................. | 18 | 2 | 12 | 0.08 | 1 | 0 | 0 | 0 | 0 | 1 |
| ......................TCCATCACCGCCGCCGCCGCCA.................................................................................................................................................. | 22 | 2 | 13 | 0.08 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...................CTCGCCACCACCGCCGCCG........................................................................................................................................................ | 19 | 1 | 13 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 |
| .......................CCACCACCGCCGCCGCTGGC................................................................................................................................................... | 20 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..............................................................................................................................................................AGCGATCCTGGCGCACCG.............. | 18 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droAna3 | scaffold_13334:780847-781036 + | dan_212 | GCGGCACGCCCAC---------------------GGCACACAGCGGT---------GGTGGCGGCG---------------------GCGGCGG-------------------------------TGGAAGCGGTG---GAGGAGGAGGT------------------GGAGG---TGGAGG---AAA------TGGTGG----------CG-----------------------------------------------------------------------------------------------------ACC---AGCCC---ATGGGCAACGCCCTGCCCCGCCTGGCCTCCCATCCCAATGCCAGC----------------------------AGCAGCAA----CTCCTCGGTGTCG--GGAGGACCG-----------------------------------CTTGGCGCCGCTGCCGTA-------GT |
| droBip1 | scf7180000396733:412830-413016 + | GCGGCACGCCCAC---------------------GGCACACAGCAGC---------AGTGGCG------------------------GCGGCGG-------------------------------------CGGTGGTGG------------AGGAGGAGGAGGAGGTGG------TGGTG---------GCGGTGGCGG----------TG-----------------------------------------------------------------------------------------------------ACC---AGCCT---ATGGGCAATGCCCTGCCCCGCCTGGCCTCCCATCCCAATGCCAGC----------------------------AGCAGCAA----TTCCTCCGTGTCT--GGAGGACCA-----------------------------------CTTGGCGCCGCAGCCGTG-------GT | |
| droKik1 | scf7180000302592:719096-719299 + | GCGGTGG------------------------------------CGGC---------GGTGCGG---------GTAACTTCAATGCTGGTGGAGC----------------------CGGCGGCACTGGAGGCGGCGTTGT------CGGTGTGCCTGGAGGAGCT---GC------------------------------------------------CGGTCCC---------G--TC----------------------------------------------------------------GTCGGTGGCGACCAGTACTCG---ATGGCCAATGCAGCAGCG-GCA--------------------------GCGGCTGCCTCCAATATGCTCCAGCAGCAGCAG----------GGCGTTG--GTGTGGGCG------------------------------------------T-TAAGC-CCGGTCCCGGCC | |
| droFic1 | scf7180000453800:737872-738058 - | GCGAAC-G---------------------------CAGCTCGGCGGC---------AGCAGCGGCG---------------------GCGGCGGCAGCAGCAGCGGTAGCCGCAGCCGGCGG------------------------------------------------------TAATGT---GAA------T--------------------------------------------------------------------------GCGGC-TGCCGTCGCCTTGGGGCTGACCACGCCCACCGGCGGCGAACGATCTCCGAGCGTGGGCAG------------------------------TGCC-----------------------------------------AGCGCAGCGGCGGCA--GCAGCGGCG-------------------------------G----TTGCTGCAGCGGTTGCA-------GC | |
| droEle1 | scf7180000491194:837912-838101 + | -CGCCATGGCCGA---------------------GGTGATCAGCGGT---------GGAGCAG------------------------GCGGCGG-------------------------------------CGGCAGCGG------------------------------------CAGCGG---AGG------CAG------------------------------TGGCGCAG--TTGGAAGCGGCAGTGGCGGCGGC-------------------------------------------------------------AGC---GGGGGCACCGCTCCGCCCAACCTGCGC-ACTACCGAGTGGCCAGT----------------------------GACAG---CAAGCTTCTCGACATCG--GCC--AGCA-------------------------------ATCATTCGCAGACTCAGTC-GA-------GT | |
| droRho1 | scf7180000778257:128891-129049 + | CGG-CGGG--------------------------------CAGCGGT---------GGTGGTG------------------------GTGGTGG-------------------------------------AGGTGGTGG------------CGG---------------------AGGAGG---AGGAGGTGGCAGCGG----------TG----------------------------------------------CCGGCAGCGGTACCGCCAG-CGGCTCCCTGTCGCCGTCCACGTC----------------------------------------------------------CGCAGCGGCAGC----------------------------AGC---------------------A--GCGGCGGCGG----------------------CAGCGGCGG----CCGCTGC-ACAATCGCA-------GT | |
| droBia1 | scf7180000302421:1236644-1236798 + | GCGGCACGCCCAC---------------------CACCCACGGCGGC---------GGACCGG------------------------GCGGAGG-------------------------------------CGGTCCCGG------------------------------------TGGCGG---CGGCGGTGGTAGCAC----------CG-----------------------------------------------------------------------------------------------------ATC---AGCCC---ATGAGCAACGCCCTGCCCCGTTTGGCCTCACACCCGAATGCC-----------------------------------------AACTCCTCCGCTTCT--GGAGCGACA--------------------------------------GGCGC--------AG-------GT | |
| droTak1 | scf7180000415872:196615-196833 - | GCAGCAGTTCCCC---------------------GGCG-GCGGTGGC---------GGCGGCGGCGGAGCGGGCAACTTTAATAC----------------------------GGTGGG------------CGGCGCTGG------------------------------------AGGCGC---CGGCGTC-GTTGGAG----------TT-----------------------------------------------------------------------------------------------------CCC---GGCGG---A--GCGAATGCCGTGCCCGGTGGCGATCAGTACTCGATGGCCAATGCGGCGGCAGCCTCCAATATGCTGCAACAGCAGCAG----GGCGTTGGAGTCG--G-CGTGGGA-------------------------------G----TGGGCGTGGGAGTCGGA-------GT | |
| droEug1 | scf7180000409271:24905-25030 - | GCGGCACGCCCAC---------------------CACACACGGCGGT---------GGACCGG------------------------GCAGTGG-------------------------------------TGGTCCAG---------------------------------------------------GTGGTAGCAG----------CG-----------------------------------------------------------------------------------------------------ACC---AGCCA---ATGGGCAATGCCCTGCCGCGTTTGGCCTCCCATCCGAATGCG-----------------------------------------AGTTCCTCCGCTTCG--G---------------------------------------------------------------------- | |
| dm3 | chrX:16966046-16966191 - | GCGGCACGCCCAC---------------------CACACACGGCGGC---------GGTCCGG------------------------GCGGAGG-------------------------------------CGGTCCCGG------------------------------------AGGCGG---CGGCGGTAGCAACAG----------CG-----------------------------------------------------------------------------------------------------ATC---AGCCA---ATGGGCAATGCCCTGCCTCGTTTGTCCTCCCACCCAACTGCC-----------------------------------------AACTCCTCCGCATCC--GTGGCGACG-------------------------------------------------------------- | |
| droSim2 | x:16083799-16083944 - | GCGGCACGCCCAC---------------------CACACACGGCGGC---------GGTCCGG------------------------GCGGAGG-------------------------------------CGGTCCCGG------------------------------------AGGCGG---CGGCGGTAGCAACAG----------CG-----------------------------------------------------------------------------------------------------ATC---AGCCA---ATGGGCAATGCCCTGCCCCGTTTGTCCTCCCACCCAACTGCC-----------------------------------------AACTCCTCCGCATCC--GTGGCGACG-------------------------------------------------------------- | |
| droSec2 | scaffold_17:799403-799548 - | GCGGCACGCCCAC---------------------CACACACGGCGGC---------GGTCCGG------------------------GCGGAGG-------------------------------------CGGTCCCGG------------------------------------AGGCGG---CGGCGGTAGCAACAG----------CG-----------------------------------------------------------------------------------------------------ATC---AGCCA---ATGGGCAATGCCCTGCCCCGTTTGTCCTCCCACCCAACTGCC-----------------------------------------AACTCCTCCGCATCC--GTGGCGACG-------------------------------------------------------------- | |
| droYak3 | X:15577820-15577968 - | GCGGCACGCCCAC---------------------AACACACGGAGGC---------GGTCCTG------------------------GCGGAGG-------------------------------------CGGAGGCAG------------------------------------TGGCGGCGGTGCCGGTAGCAACAG----------CG-----------------------------------------------------------------------------------------------------ATC---AGCCA---ATGGGCAATGCCCTGCCCCGTTTGGCCTCCCACCCGACTGCC-----------------------------------------AACTCCTCCGCATCC--GTGGCGACG-------------------------------------------------------------- | |
| droEre2 | scaffold_4690:7319307-7319449 - | GCGGCACGCCCAC---------------------TACACACGGCGGC---------GGTCCGG------------------------GCGGAGG-------------------------------------CGGTCCCGG------------------------------------AGGCGC---CGGC---AGCAACAG----------CG-----------------------------------------------------------------------------------------------------ACC---AGCCA---ATGGGCAATGCCCTGCCACGTTTGGCCTCCCACCCGACTGCC-----------------------------------------AATTCCTCCGCATCC--GTGGCGACG-------------------------------------------------------------- | |
| dp5 | XL_group1e:1110993-1111167 - | GCGGCACCCCCAC---------------------CACGCATGGCGGCACCGGGATGAGTGGAG------------------------GAGGAGG-------------------------------------CGGCGGCGG------------------------------------TGGCGG---CAACGGCAGCAA------------------------------TGCCGCAA--TG--------------------G-----------------------------------------------------ATC---AACCC---ATGGGTAATGCACTGCCTCGCCTGGCGTCACATCCAAATACCG--------------------------------------GGAGCTCCTCGGGAGCT--GGAGGAGCGC--------------------------------------------CAATGGGA-------GG | |
| droPer2 | scaffold_30:84935-85106 + | GCGGCACCCCCAC---------------------CACGCATGGCGGCACCGGGATGAGTGGAG------------------------GAGGAGG-------------------------------------CGGCGGCGG------------------------------------TGGCGG---CAACGGCAGCAA------------------------------TGCCGCAA--TG--------------------G-----------------------------------------------------ATC---AACCC---ATGGGTAATGCACTGCCTCGCCTGGCGTCACATCCAAATACCG--------------------------------------GGAGCTCCTCGGGAGCT--GGAGGAGCGCC-----------------------------------------------AATG-------GG | |
| droWil2 | scf2_1100000004768:1760742-1760952 + | dwi_2111 | ACGGAACTCACCCAGCGCCAATGGTGCTGGCGGCGG----CGGCGGCGGT------GGAGG----------------------------------------------------------------------AGGTG---GAGGTGGAGGC------------------GGCAGTGGAGGAGC---A---------GG---------------------AGGTGCC---------G--CTGCCAATGGTAATGGCGGCGGT---A-----ATGGC-AACAATA-------------------------------GCC---ACGAA---AGCAGCAACAATTCGCCCAA-----C-----------------------------------------------------------------AGTATTGCCGGAGGACCT---------------GGAGGTGGCAGCGGCGC----CGGCGGTAGCAATAATA-------AT |
| droVir3 | scaffold_12970:9691838-9692021 - | GCGGCACACCCAC---------------------CACACATGGCGGC---------AGCGTGG------------------------GAGGTGG-------------------------------------CA-------------------------------------------------------------------TGCAGTCTAGCGAACAGTCGTCTCTGATGCCACAGGGC--------GTAATGGCGGG---------------------------------------------------------------CGCA---TTGGGCAATGCTCTGCCACGACTGGCCTCTCATCCAAATCCC-----------------------------------------AGCGCAGTGGT-----------TACTCCCATGGGGGCG-----------------------CATTACGGTTCCGTCGGA-------GT | |
| droMoj3 | scaffold_6473:11876800-11876987 + | GCGGCACACCCAC---------------------CACGCATGGTGGC---------AGCGTGG------------------------GCGGCGG-------------------------------------CA-------------------------------------------------------------------TGCAGTCTAGTGAACAGTCGTCTCTGATGCCACAGGGTG--------------------G---------------------------------------------------------------A---TTGTGTAATGCACTGCCTCGACTGGCCTCCCATCCAAATACC-----------------------------------------GGCGCAGTCGT-----------TACGCCCATTGGTGCGCATTATGGTGCCATCGGT-G----TGGGCGG--CACTGGCG-------GC | |
| droGri2 | scaffold_14853:346595-346782 + | GCGGCACGCCCAC---------------------AACACATGGTGGC---------AGCGGCGGAG---------------------GCGGAGG-------------------------------------CGGCGTTGG------------------------------------TGGTGG---CGGTGGTGGTG----TTCAGTCCAACGAACAGTCGTCAATGATGCCACAGGG-----------------------------------------------------------------------------------GGCA---TTAAGCAATGCACTGCCACGACTGGCCTCCCACCCAAACTCC-----------------------------------------------AGCGCA-----GTGGTTACGCCAA--------------------------------TGGGCGTCGCAG--GTG-------CT |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/15/2015 at 02:58 PM