ID:dan_2075 |
Coordinate:scaffold_13266:17263473-17263623 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
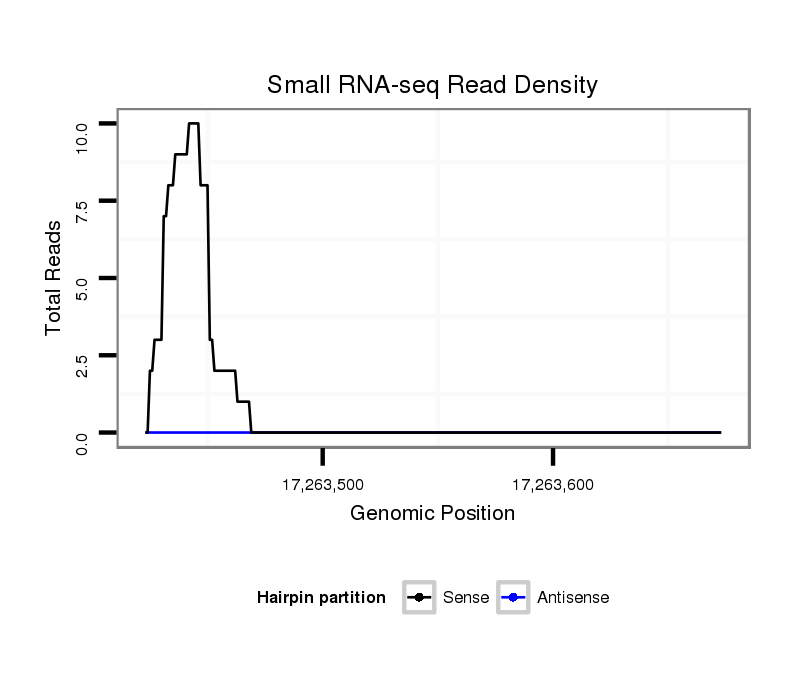
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
CDS [Dana\GF13729-cds]; exon [dana_GLEANR_13736:1]; intron [Dana\GF13729-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GGATCAGAGTACCGATGATCAGGTTAGCGTCAACTCACAGGATGCCGATGGTGAGTGCTTTTCGCTTTAGTCCTTTGTTTCAGGGTGCTGTCTCCCCTCTGGTTCGAGGTTCGAGGATGAAGGAAGAAATTCGATTGCGTGTCCTGTCATCTTTGGGTTCTTGGGGCCAAGTAGTTGGCATTTTCTAGCTGCCGCGTGTTGTAAAATTCTAGCTAGAGTTATTTTTAGTCTCCTAGTTTTTTCCTTTTATT **************************************************(((((.....)))))..((((.(((((...))))).)))).........((((((.((((((((((((((((((.......((....))..)))).)))))))))))..))).)))))).((((((((......)))))))).........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M044 female body |
V105 male body |
V055 head |
V106 head |
M058 embryo |
V039 embryo |
|---|---|---|---|---|---|---|---|---|---|---|---|
| .....................................................................................................................................................TCTTTGGGTTCTGGGGGGA................................................................................... | 19 | 3 | 20 | 43.00 | 860 | 583 | 223 | 20 | 34 | 0 | 0 |
| .....................................................................................................................................................TCTTTGGGTTCTGGGGGG.................................................................................... | 18 | 2 | 12 | 20.83 | 250 | 180 | 39 | 3 | 28 | 0 | 0 |
| ....................................................................................................................................................GTCTTTGGGTTCTGGGGG..................................................................................... | 18 | 2 | 15 | 18.53 | 278 | 165 | 86 | 8 | 12 | 6 | 1 |
| ......................................................................................................................................................CTTTGGGTTCTGGGGGG.................................................................................... | 17 | 2 | 20 | 3.55 | 71 | 0 | 0 | 71 | 0 | 0 | 0 |
| ......................................................................................................................................................CTTTGGGTTCTGGGGGGC................................................................................... | 18 | 2 | 8 | 3.38 | 27 | 2 | 5 | 16 | 4 | 0 | 0 |
| ........GTACCGATGATCAGGTTAGC............................................................................................................................................................................................................................... | 20 | 0 | 1 | 3.00 | 3 | 0 | 1 | 1 | 0 | 1 | 0 |
| ....................AGGTTAGTGTAAACTCTCAG................................................................................................................................................................................................................... | 20 | 3 | 3 | 1.67 | 5 | 5 | 0 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................ATCTTTGGGTTCTGGGGGGA................................................................................... | 20 | 3 | 12 | 1.50 | 18 | 0 | 13 | 0 | 5 | 0 | 0 |
| ....................................................................................................................................................ATCTTTGGGTTCTGGGGGG.................................................................................... | 19 | 2 | 3 | 1.33 | 4 | 0 | 3 | 0 | 1 | 0 | 0 |
| .............GATGATCAGGTTAGCGTCAACTCACAG................................................................................................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ........GGACCGATGATCAGGTTAGC............................................................................................................................................................................................................................... | 20 | 1 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................................................................................TTGGGTTCTTGGGCCCA.................................................................................. | 17 | 1 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........GTACCGATGATCAGGTTAGCGT............................................................................................................................................................................................................................. | 22 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................CAGGTTAGCGTCAACTCACAGGATGCC............................................................................................................................................................................................................. | 27 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........ACCGATGATCAGGTTAGC............................................................................................................................................................................................................................... | 18 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ..........................................................................................................................................................................GAAGTTGGCATTTTCTAGTTGCA.......................................................... | 23 | 3 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ......................................................................................................................................................CTTTGGGTTCTGGGGGCCGTGT............................................................................... | 22 | 3 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ..ATCAGAGTACCGATGATCAGGT................................................................................................................................................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ....CAGAGTACCGATGATCAGGT................................................................................................................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ....................................................................................................................................................GTCTTTGGGTTCTTGGGG..................................................................................... | 18 | 1 | 2 | 1.00 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| ..ATCAGAGTACCGATGATCAGGTTAGC............................................................................................................................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .......................................................................................................................................................TTTGGGTTCTCGGGGCCA.................................................................................. | 18 | 1 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................TTTGGGTTCTGGGGGCCATCT............................................................................... | 21 | 3 | 2 | 0.50 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................GTCTTTGGGTTCTGGGGGGCA.................................................................................. | 21 | 3 | 4 | 0.50 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................CTTTGGGTTCTGGGGGCA................................................................................... | 18 | 2 | 11 | 0.45 | 5 | 2 | 2 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................TTTGGGTTCTTGGGGGGGAGT............................................................................... | 21 | 3 | 11 | 0.45 | 5 | 1 | 3 | 0 | 1 | 0 | 0 |
| .....................................................................................................................................................TCTTTGGGTTCTGGGGG..................................................................................... | 17 | 1 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................TTTGGGTTCTTGGGGGGGAGTA.............................................................................. | 22 | 3 | 3 | 0.33 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................CTTTGGGTTCTGGGGGCCGT................................................................................. | 20 | 3 | 10 | 0.30 | 3 | 0 | 0 | 3 | 0 | 0 | 0 |
| ....................................................................................................................................................ATCTTTGGGTTCTGGGGGGAA.................................................................................. | 21 | 3 | 8 | 0.25 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................TCTTTGGGTTCTTGGGGGA................................................................................... | 19 | 2 | 12 | 0.25 | 3 | 1 | 1 | 0 | 1 | 0 | 0 |
| .......................................................................................................................................................TTTGGGTTCTTGGGGGC................................................................................... | 17 | 1 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................GTCTTTGGGTTCTAGGGG..................................................................................... | 18 | 2 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................CTTTGGGTTCTTGGGGAG................................................................................... | 18 | 2 | 14 | 0.14 | 2 | 0 | 2 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................CTTTGGGTTCTGGGGGACGA................................................................................. | 20 | 3 | 8 | 0.13 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................TTTGGGTTCTGGGGGCCG.................................................................................. | 18 | 2 | 8 | 0.13 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................TTTGGGTTCTTGGGGCCTT................................................................................. | 19 | 2 | 8 | 0.13 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................GTCTTTGGGTTCTTGGGGG.................................................................................... | 19 | 2 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................CTTTGGGTTCTGGGGGGCATG................................................................................ | 21 | 3 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................CTTTGGGTTCTCGGGGCCGT................................................................................. | 20 | 3 | 9 | 0.11 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................................TTGGGTTCTTGGGGGGGAGTA.............................................................................. | 21 | 3 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................CTTTGGGTTCTGGGGGTC................................................................................... | 18 | 2 | 19 | 0.11 | 2 | 0 | 0 | 2 | 0 | 0 | 0 |
| .......................................................................................................................................................TTTGGGTTCTTGGGGGCG.................................................................................. | 18 | 2 | 20 | 0.10 | 2 | 0 | 0 | 1 | 1 | 0 | 0 |
| ........................................................................................................................................................TTGGGTTCTTGGGGGC................................................................................... | 16 | 1 | 11 | 0.09 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................ATTTGGGTTCTGGGGGCC................................................................................... | 18 | 2 | 13 | 0.08 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................TTTGGGTTCTTGGGGGCGTGT............................................................................... | 21 | 3 | 13 | 0.08 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................CGTCTTTGGGTTCTGGGGGG.................................................................................... | 20 | 3 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................AATCTTTGGGTTCTGGGGGG.................................................................................... | 20 | 3 | 14 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................CTTTGGGTTCTCGGGGACG.................................................................................. | 19 | 3 | 14 | 0.07 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ........................................................................................................................................................TTGGGTTCTTGGGGCCGTCT............................................................................... | 20 | 3 | 14 | 0.07 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................CTTTGGGTTCTTTGGGGCGA................................................................................. | 20 | 3 | 18 | 0.06 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| ....................................................................................................................................................GTCTTTGGGTTCTGGGGGCA................................................................................... | 20 | 3 | 19 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................TTGGGTTCTGGGGGCCT.................................................................................. | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................TCTTTGGGTTCTGGGGGGAA.................................................................................. | 20 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................CTTTGGGTTCTGGGGGACG.................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
| .......................................................................................................................................................TTTGGGTTCTTGGGGGGA.................................................................................. | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 | 0 |
|
CCTAGTCTCATGGCTACTAGTCCAATCGCAGTTGAGTGTCCTACGGCTACCACTCACGAAAAGCGAAATCAGGAAACAAAGTCCCACGACAGAGGGGAGACCAAGCTCCAAGCTCCTACTTCCTTCTTTAAGCTAACGCACAGGACAGTAGAAACCCAAGAACCCCGGTTCATCAACCGTAAAAGATCGACGGCGCACAACATTTTAAGATCGATCTCAATAAAAATCAGAGGATCAAAAAAGGAAAATAA
**************************************************(((((.....)))))..((((.(((((...))))).)))).........((((((.((((((((((((((((((.......((....))..)))).)))))))))))..))).)))))).((((((((......)))))))).........************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M058 embryo |
M044 female body |
V055 head |
V106 head |
V039 embryo |
|---|---|---|---|---|---|---|---|---|---|---|
| ...................................................................................................................................................................................................................................AAGAGGATCAAAACAGGAAAATA. | 23 | 2 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................................................................................................GAGGACAGTAGAAACCCGAGC.......................................................................................... | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................CCAATCAGAGGATCAAAAAA......... | 20 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................GGGGATACCAAGCTCCAACA.......................................................................................................................................... | 20 | 3 | 3 | 0.33 | 1 | 0 | 0 | 1 | 0 | 0 |
| .......................................................ACCAAAAGAGAAAGCAGGAAACA............................................................................................................................................................................. | 23 | 3 | 3 | 0.33 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................................CCACGCACGAAAAGAGAACT...................................................................................................................................................................................... | 20 | 3 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................AGAGGATCAAAAAGCGAAAAGAA | 23 | 3 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................CAGCGAAAGCAGGAAACTAAG.......................................................................................................................................................................... | 21 | 3 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................AAAAATCAGAGGCACAAAA........... | 19 | 2 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................................................AAAAAACAGAGGACCAAAAAA......... | 21 | 2 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................AAGGGGCACAACATTTTA............................................ | 18 | 2 | 9 | 0.11 | 1 | 0 | 0 | 0 | 1 | 0 |
| .................................................CCACTAACGAACAGCGAAAA...................................................................................................................................................................................... | 20 | 3 | 12 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 |
| ................................................ACCACCCACGAAAAGCAA......................................................................................................................................................................................... | 18 | 2 | 14 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................................CGAAGATCAAAAAAGGAAA.... | 19 | 2 | 17 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 |
| ...........................................CCGCAACCACTCACGAAAAA............................................................................................................................................................................................ | 20 | 3 | 18 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droAna3 | scaffold_13266:17263423-17263673 + | dan_2075 | GGATCAGAGTACCGATGATCAGGTTAGCGTCAACTCACAGGATGCCGATGGTGAGTGCT-T--TTCGCTT-TAGTC----------------------------------------------CTT-------------------T---GTTTCAGGGTGCTGTCTCCCCTCTGGTTCGAGGTTCGAGGATGAAGGAAGAAA--------TTCGATTGCGTGTCCTGTCATCTTTGGGTTCTTG-GGGCCAAGTA-----------------------------------------------------GTTGGCATTTTCTAG---------------------CTGCCGCGTG--TTGTAAAATTC-------TAGCTAGAGTTATTTTTAGT---------------------------CT------------C-C------------------TA--GTTTTTT-CCTTTT-----------------ATT |
| droBip1 | scf7180000396735:531254-531467 + | GGACCAGAGTACCGATGACCAGGCTAGCGTCAACTCACAGGATGCCGATGGTGAGTGG-----------------------------------------------------CTTTCAGAGTCCGGGG------GCC--------------------------------------------------TTC----CA-AGAAAT-------ATCGATTGCGTGTCCTGTCATCTTTTTGTTCTTCTGGC-CAACTA-----------------------------------------------------GTTTGCATTTTCTAG---------------------CTGCTGCGTG--TTGTAAAATTC-------TTGCTAGAGTTATTTATAGT---------------------------TT----------TCC----------------C----ATATTTTTTCGCCTTTT-----------------ATT | |
| droKik1 | scf7180000302682:1563543-1563756 + | GGAGCAGAGCACCGATGACCAGCTGAGCGTCAACTCACAAGAAGCCGATGGTGAGTGGAGT--TG-GTAT-TATTAACTTCA---------------GGAGTTCCTTTAGTC----------CCT-----------------------------------------------------------------------TG-TT--------TTTGTTGGCATGTCCTGTCAACTTCTTTT---C--ATCA---------------------------------------------TATTTTTTCTTCG-TTTTACATTT----G---------------------CCACTGCGTG--TTGTAAAATTC-------TTGGGTCAGATATTTTTAGT---------------------------TT----------T---C-------------------C--------T-TCCCGT-----------------ACT | |
| droFic1 | scf7180000453948:643810-644089 + | AGATAATAGTACCGATGACCAGCTGAGCGTGAACTCACAAGATGCCGATGGTAAGTGGAGT--TTTAAAG-TCTTG----CGAGTTCTT------CAACTGCCTGCTTAAACTGTCGGAATCCCATG------GTTTG------G----------------------------------------TGCC----CAAAGTTA--------TTCGATGGCGTGTCCTGTCAAGTTTTTCC---T--GCCTCAACTG--------------C--CATCCTCGTGACTTTTT-----TCCAGCTCTCTAG-CTTTGCATTTCTT-GCCT------------------CTGCTGCGTG--TTGCAAAATAC-------TTGCCTCGGATATTTTTAC---------------------------TCT----------TTT----------------C----ACG-------------TACTTTT-----------ACT | |
| droEle1 | scf7180000491107:1198817-1199102 + | AGACATAAGTACCGATGACCAACTGAGCGTCAACTCACAGGATGCCGATGGTAAGTGGAGT--TTCGTTT-TATTT----GA---------------ACTGCTTCTTTAAGCCATCGGAATGCGATG------GTTTT------T----------------------------------------CATT----CAAAGATTT-------TCCGATTGCGTGTCCTGTCAACTTTTTCT---T--GGCTTAAATG--------------C--CAGAC--TGTT-TTTTC-----TGCAGCTCCTTCG-TTTTGCATTTGCT-GCATTTGCCGCTGGT-----ACGAGGTGCGTG--CTGTAGAATTC-------TTGCCAGGGATATTTTCACT---------------------------TC------------C-C-------------------CTG-------------CACTTTTTCTGTGCTT----- | |
| droRho1 | scf7180000777200:78521-78740 - | AGATATAAGTACCGATGACCAGCTGAGCGTCAACTCACAGGATGCCGATGGTAAGTGGAGT--TTTATTT-TATTT----GA---------------ACGGCT-----------------------G------GTT--------TTTTATTT--------------------------------A-ATC----CAAAGACA--------TTCGATTGCGTGTCCTGTCAACTTTTTCT---T--GGCCCAACTG--------------CC-CCGAC----------------------------CT-TTTTGCATTTTC-------------------------CGCTGCGTG--TTGTAGAATTC-------TTGCCTGGGATATTTTTAC---------------------------TTT----------TTC----------------C----C------------CTGC-----------------ACT | |
| droBia1 | scf7180000301506:805531-805797 + | GGATCACAGTACCGATGACCAGCTGAGCGTCAACTCACAGGATGCCGATGGTAAGTGGAGT--TTTATGT-TGTTT----CT---------------GCTGCTTCTTAAGTCTACAGGAATCCCAAGT-----TGTTT------TTGC----------------------------TTACGTTCTCTAC----CCAAGTTA--------TTCCATTGCGTGTCCTGTCAACTTTTTCT---T--GGCTTAACTG--------------C--C--------------------TTAAATCTGCTTCGTTTTTGCATTTTCC-GCTTTTG---------------GCGCTGCGTG--TTGGAAAATTC-------TTGCCTGGGAGATTGGTAC---------------------------TTT----------TTT----------------C----ACG-------------T-----------------ACT | |
| droTak1 | scf7180000415870:357139-357413 + | GGATCAAAGTACCGATGACCAACTGAGCGTCAACTCACAAGATGCCGATGGTAAGTGGAGT--TTTATTT-TATTT----GA---------------ACTGCTTCATAAAGCTTCCGGAATCCAATG------GGTTT------T----------------------------------------TTTC----CAAAGATA--------TTCGATTGCGTGTCCTGTCAACTTTTCCT---T--GGCTCAACTG--------------C--CAGTC--GTTT-TTTTTTACCTAAAATCTCCTTAG-CATTGCATTTGCT-T---------------------CTGCTGCGTG--TTGAAAAATTC-------TTGCCTAGGATTCTG----------TGTAC----------------TTT----------TTT----------------C----ACG-------------TACTTTTTCCT-------AGT | |
| droEug1 | scf7180000409209:285974-286266 + | CGATCAAAGTACCGATGACCAACTGAGCGTCAACTCACAGGATGCCGATGGTAAGTGCAGT--TTTATTTCTGTTC----CT---------------TTCCCCTTTTTCAACTTCAGGAATGTTTTG-TACTGG----------------------------------------------------CTC----TAAAGATA--------TTCGATTGCGTGTTCTGTCAACTTTTTCT---T--GGCTTAACTG--------------C--CAGGC----------------TTAAATCTCCTTCG-TTTTACATTTTCC-ACTGCTGCTC---CTCCTGCCTCTGCTGCGTG--TTGCAAAATTC-------TTGCCTGGGATATTTTTACTTTTTGTGTACGTTTCTGTTTCTGTTTTTT----------TTTCA-----------------------------------------------------ACT | |
| dm3 | chr2R:5892404-5892711 - | GGATCAAAGCACCGATGACCAGCTGAGCGTCAACTCACAGGATGCCGATGGTAAGTGGAGTGTTCTACTT-TATTC----GA---------------GCAGCTTCTTTAAGCTGCCGGAATCCGTTG------TGCTCCTCTT-C----------------------TTTTTTTTTCTATTTTCGTTTA----CAAAGCAA--------TTCGATTGCGTGTCCTGTCAACTTTTTCT---T--GGCTTAACTG--------------C--CAGCC----------------CAAAGTCCTCTACA-TTTTGCATTTTCT-G---------------------CTGCTGCGTG--TTGGAAAATTC-------CTGCCGGGGATAGTTAAAAAG---------------------------------------TGCAACTACATACACATACACATA--CT--------------TTTTTCAGAGCTCTTTTC | |
| droSim2 | 2r:6668731-6669017 - | dsi_12035 | GGATCAAAGCACCGATGACCAGCTGAGCGTCAACTCACAGGATGCCGATGGTAAGTGGAGT--TTTACTT-TATTC----GA---------------GCAGCTTCTTTAAGCTGCCGGAATCCGGTG------TGCTCCTCTTTC----------------------TTTTTTGTGCTATTTTCGTTTC----CAAAGCAA--------TTCGATTGCGTGTCCTGTCAACTTTTTCT---T--GGCTTAACTG--------------C--CAGCC----------------GTAAGTCCTCTTCG-TTTTGCATTTTCT-G---------------------CTGCTGCGTG--TTGGAAAATTC-------CTGCCGGGGATAGTTGCAC---------------------------TTT----------TTC----------------C----ACA-------------TACTTTTCCAGAGCTC----- |
| droSec2 | scaffold_1:3506309-3506591 - | GGATCAAAGCACCGATGACCAGCTGAGCGTCAACTCACAGGATGCCGATGGTAAGTGGAGT--TTTACTT-TATTC----GA---------------GCAGCTTCTTTAAGCTGCCGGAATCCGGTG------TGCTCCTCTTTT--------------------------TTTTTCTATTTTCGTTTC----CAAAACAA--------TTCGATTGCGTGTCCTGTCAACTTTTTCT---T--GGCTTAACTG--------------C--CAGCC----------------CAAAGTCCTCTTCG-TTTTGCATTTTCT-G---------------------CTGCTGCGTG--TTGGAAAATTC-------CTGCCGGGGATAGTTGCAC---------------------------TTT----------TTC----------------C----ACA-------------TACTTTTCCAGAGCTC----- | |
| droYak3 | 2L:18521063-18521354 - | GGATCAAAGCACCGATGACCAGCTGAGCGTCAACTCACAGGATGCCGATGGTAAGTGGAGT--TTTACTT-TATTA----CAACCGCTTCTA--CGAACAGCTTCTTTAAGCTGCCGGAATCCGGTG------TGTTT------TTGTATTT--------------------------------ATTTC----CATAGCAA--------TTCGATTGCGTGTCCTGTCAACTTCTTCT---T--GGCTTAACTG--------------C--CAGCC----------------CTAAGTCTGGTTCG-TTTTGCATTTTCT-G---------------------CTGCTGCGTG--TTGCAAAATTC-------CTGCCGGGGAAAGTTGCAC---------------------------TTTTATTTTTACTTTG----------------C----ACT-------------TACTTTTTCAGTAC------- | |
| droEre2 | scaffold_4929:8476644-8476941 + | GGATCAAAGCACCGATGACCAGCTGAGCGTCAACTCACAGGATGCCGATGGTGAGTGGCGT--TTTACTT-TATTC----GAATCGC---TATTCGAACAGCTTCTTCAAGCTGCCGGAATCCGGTG------TGCTTTTCTTTTTGCATTT--------------------------------ATTTC----CATAGCAA--------CTCGATTCCGTGTCCTGTCAAGTTTTTCT---T--GGCTTAACTG--------------C--CAGCC----------------CTAAGTCTTGTTGG-TTTTGCATTTGCT-GCTGCTGCTG------------CTGCTGCGTG--TTGCAACATTC-------CTGCCGGCTAAAGTTGCAC---------------------------TTT----------TTT----------------C----ACT-------------CACTTTTTTTGAATAG----- | |
| dp5 | 3:6108737-6108900 - | GGATCACAGTCAGGATGATCAGCAGAGCCTCAACTCCCAGGATGGAGAAGGTGAGTGGGG------------------------------------------------------------------------------------------------------------------------------------------CCC--------TTCGATGGCGTGTCCTGTCATCCATTTCT---T--TGCGTGTGCGTCGGCATTCCTTT----------------------------------------------------------------------------GGGGGGGGGC--GTGCAACATTC-------TTTCTAGGTTTATTTTTAGT---------------------------TT------------C-C-----------------------------------------------------ATT | |
| droPer2 | scaffold_2:6312056-6312218 - | GGATCACAGTCAGGATGATCAGCAGAGCATCAACTCCCAGGATGGAGAAGGTGAGTGGGG------------------------------------------------------------------------------------------------------------------------------------------CCC--------TTCGATGGCGTGTCCTGTCATCCTTTTCT---T--TGCGTGTGCGTCGGCATTCCTTT-----------------------------------------------------------------------------GGGGGGGGC--TTGCAACATTC-------TTTCTAGGTTTATTTTTAGT---------------------------TT------------C-C-----------------------------------------------------ATT | |
| droWil2 | scf2_1100000004513:5317145-5317395 - | GGAACATAATACCGATGATCAGCTGAGCATTAACTCACAGGATGGTGATGGTGAGTCCTA----------------------------------CTAACTAGTTTAGTAAAT------------------------------------------------------------------------------GAGGGTAGAAAACTTGTCTTAAAATTGCGTGTCCTGTCATTGGATTTT---T--TGCAACTATTTCTGGATTTCTTTACGTCAATT----------------TTATGTTTCTTTCT-T-------------T---------------------TTGCTGCGTGTGTTGCAGTTTTTCATTTTATTTTTAGTAATAGTTGTTT--TTTGTGTGTGCTTCG----------TTT----------TT-----------------------------------CTAT-----------------ATT | |
| droVir3 | scaffold_12875:407362-407417 - | CGATAATGGGGCCGATGATAGTCAGAGCATTAATTCACAAGATGGCGACGGTAAGT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droMoj3 | scaffold_6496:5084521-5084576 + | CGATAACACGGCCGATGATAATCAAAGCGTCAACTCACAGGAAGGCGATGGTAAGT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- | |
| droGri2 | scaffold_15245:10560764-10560819 + | TGATAACGGGGCCGATGATCAGATAAGCCTCAACTCACAAGATGGCGACGGTAAGT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droKik1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droRho1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBia1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droTak1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dm3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSim2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droSec2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droYak3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| dp5 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droPer2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droWil2 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droVir3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droMoj3 |
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/18/2015 at 03:37 AM