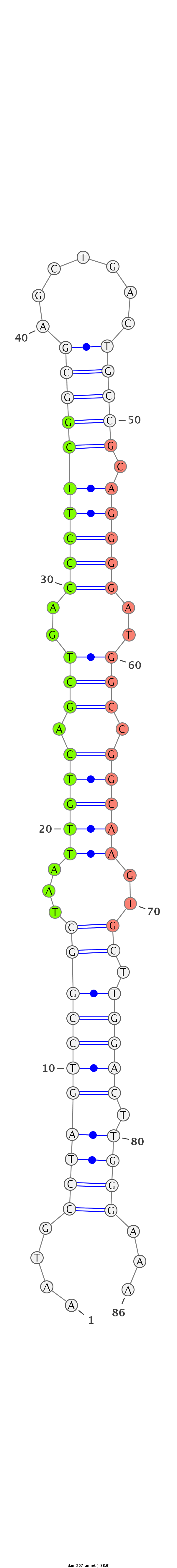
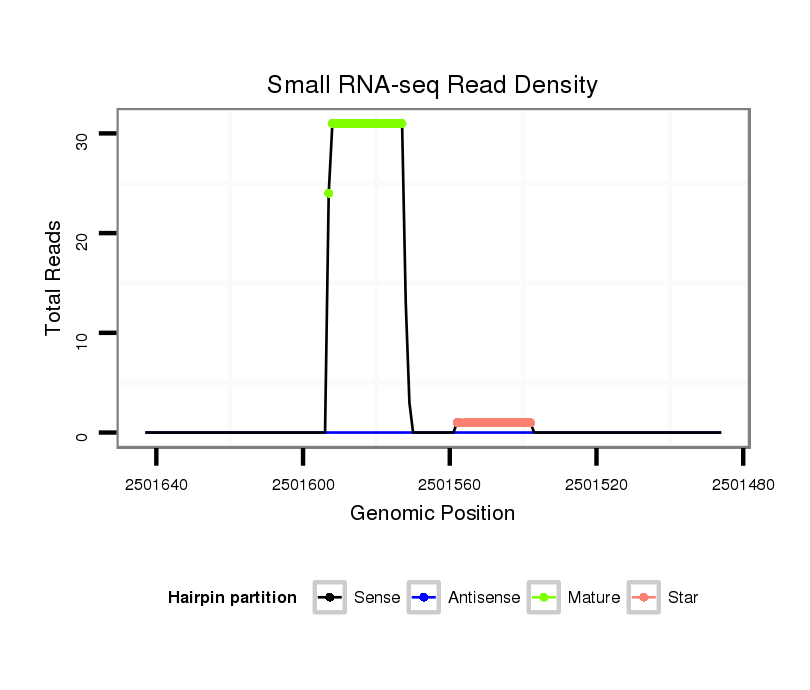
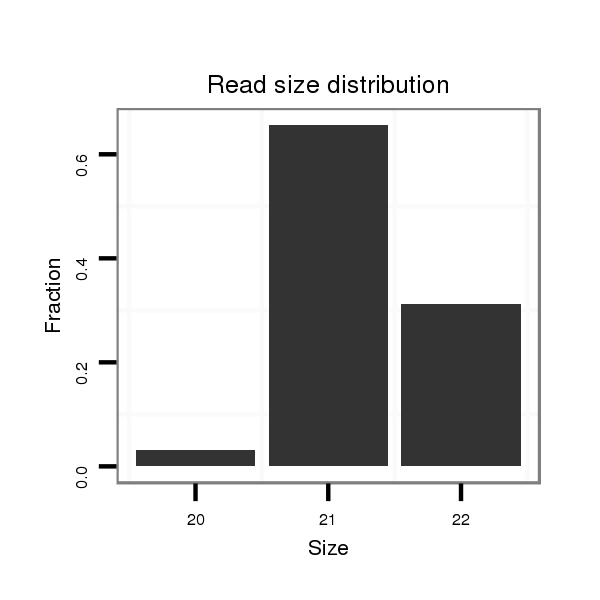
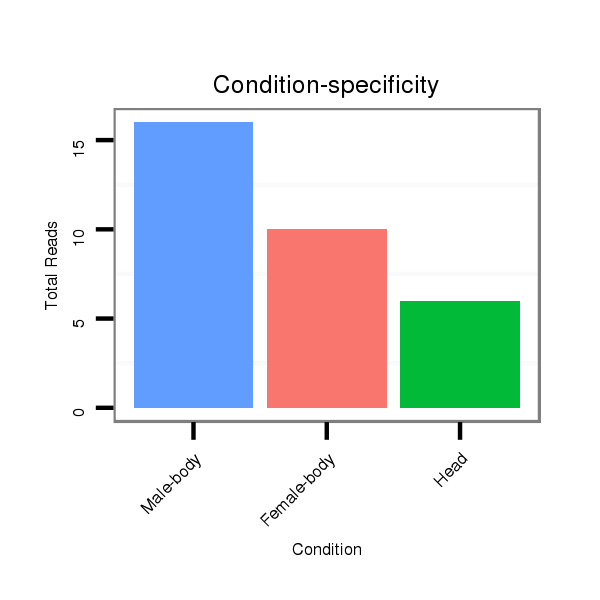
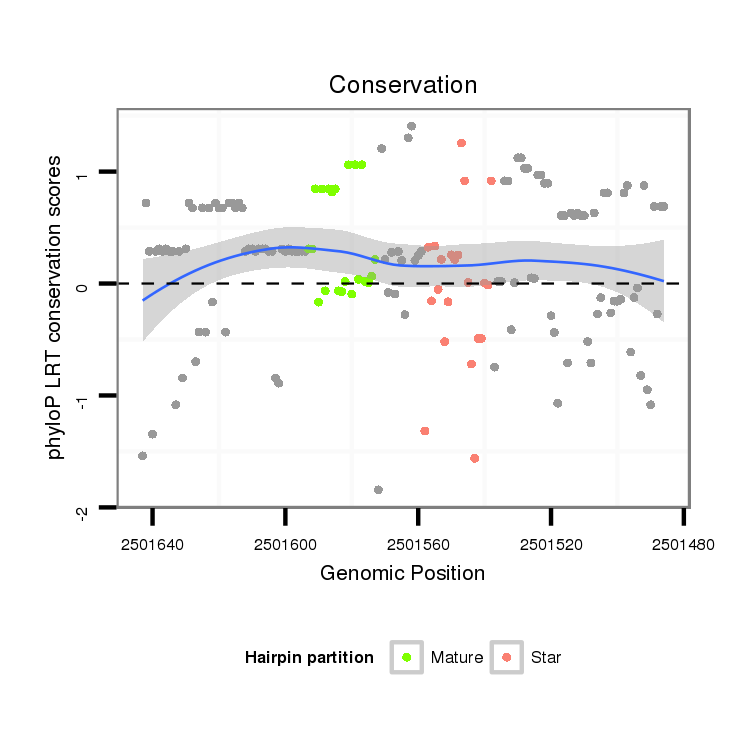
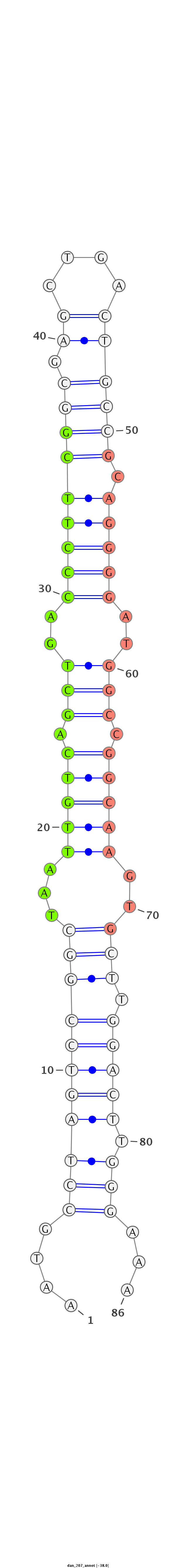
| ..................................................TAATTGTCAGCTGACCCTTCG....................................................................................... |
21 |
0 |
1 |
17.00 |
17 |
12 |
3 |
2 |
0 |
0 |
| ..................................................TAATTGTCAGCTGACCCTTCGG...................................................................................... |
22 |
0 |
1 |
7.00 |
7 |
0 |
5 |
1 |
1 |
0 |
| ...................................................AATTGTCAGCTGACCCTTCGGTT.................................................................................... |
23 |
2 |
1 |
5.00 |
5 |
2 |
1 |
2 |
0 |
0 |
| ...................................................AATTGTCAGCTGACCCTTCGG...................................................................................... |
21 |
0 |
1 |
3.00 |
3 |
1 |
1 |
1 |
0 |
0 |
| ...................................................AATTGTCAGCTGACCCTTCGGC..................................................................................... |
22 |
0 |
1 |
3.00 |
3 |
1 |
1 |
0 |
1 |
0 |
| ..................................................TAATTGTCAGCTGACCCTTCGT...................................................................................... |
22 |
1 |
1 |
2.00 |
2 |
1 |
0 |
1 |
0 |
0 |
| ..................................................TAATTGTCAGCTGACCCTTCGGTT.................................................................................... |
24 |
2 |
1 |
2.00 |
2 |
1 |
0 |
1 |
0 |
0 |
| ...................................................AATTGTCAGCTGACCCTTCG....................................................................................... |
20 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................................................TAATTGTCAGCTGACCCTTCGGCTTT.................................................................................. |
26 |
3 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
| ..................................................TAATTGTCAGCTGACCCTTCT....................................................................................... |
21 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ..................................................TAATTGTCAGCTGACCCTTCA....................................................................................... |
21 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .....................................................................................GCAGGGGATGGCCGGCAAGTGT................................................... |
22 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...................................................AATTGTCAGCTGACCCTTCGGT..................................................................................... |
22 |
1 |
1 |
1.00 |
1 |
0 |
1 |
0 |
0 |
0 |
| ..................................................TAATTGTCAGTTGACCCTTCG....................................................................................... |
21 |
1 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| .....................................................................................GCAGGGGATGGCCGGCAAGTG.................................................... |
21 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...................................................AATTGTCAGCTGACCCTTCGGCT.................................................................................... |
23 |
1 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
| .............................................................................TGACTGACGCGGGGGATGCCC............................................................ |
21 |
3 |
3 |
0.33 |
1 |
0 |
0 |
0 |
1 |
0 |
| .....................................................................................................................GAAAAGAACCCGACACCACA..................... |
20 |
3 |
11 |
0.09 |
1 |
0 |
0 |
0 |
0 |
1 |
| ................................................................................................................................AAGACCACACCGGACAACA........... |
19 |
3 |
20 |
0.05 |
1 |
0 |
0 |
0 |
0 |
1 |