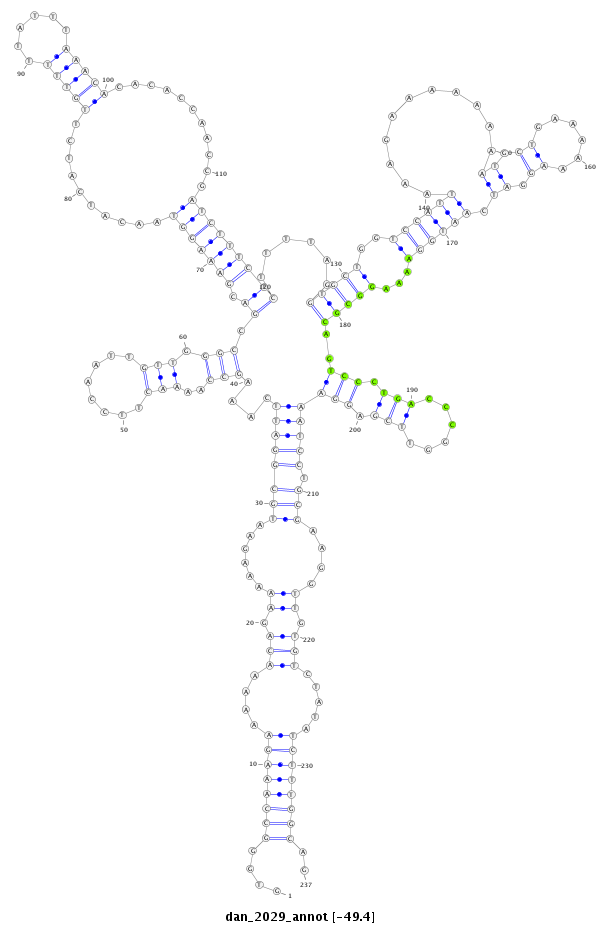
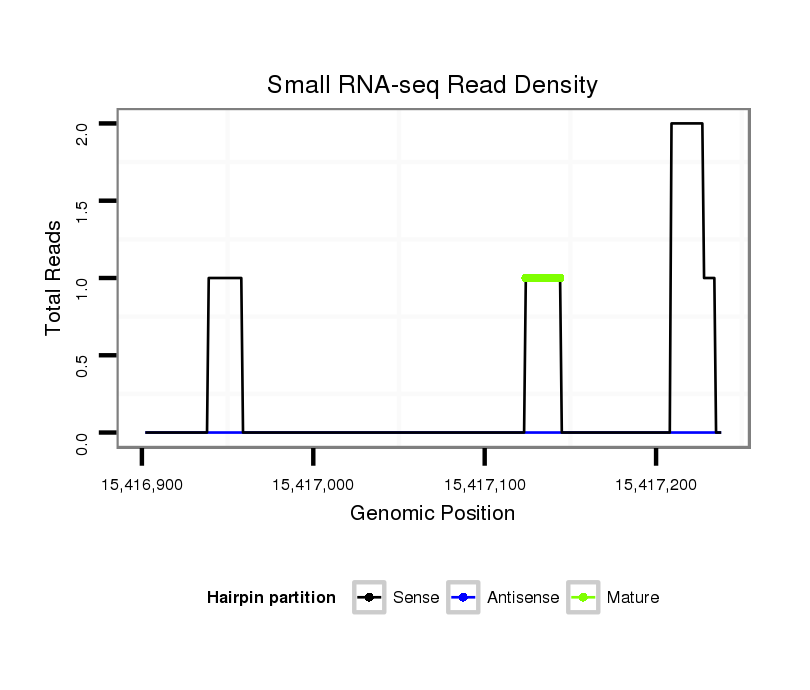
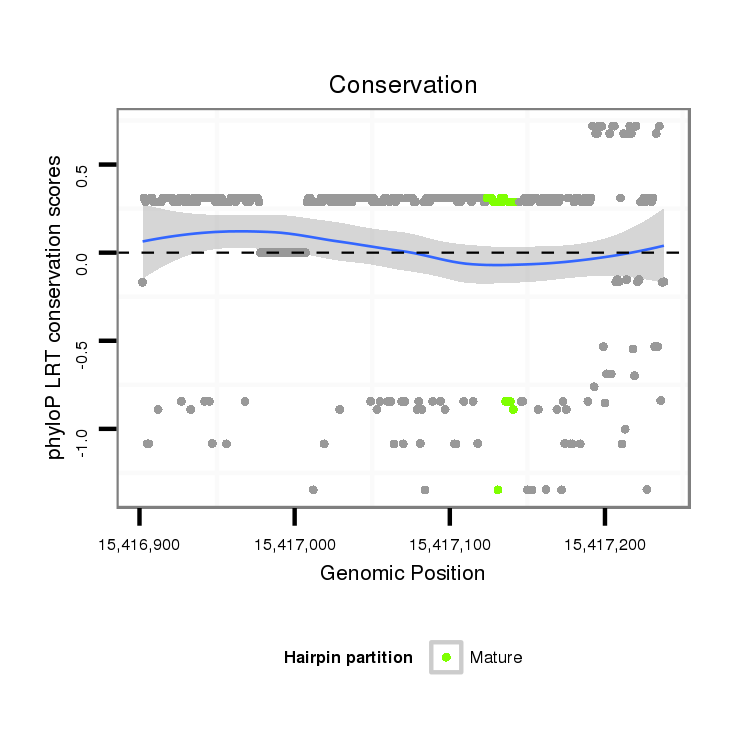
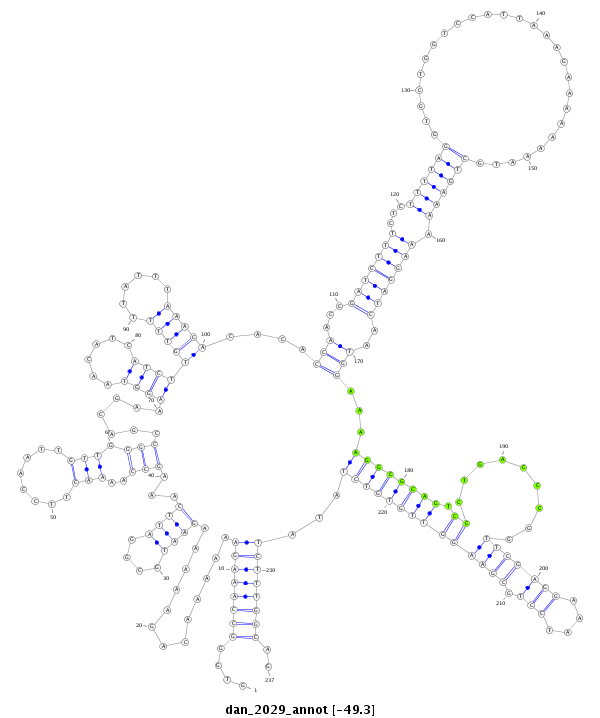
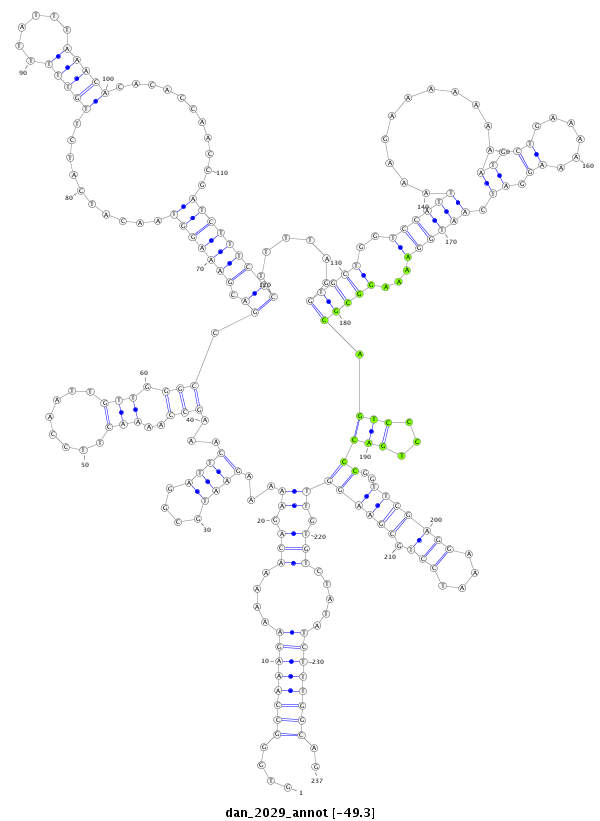
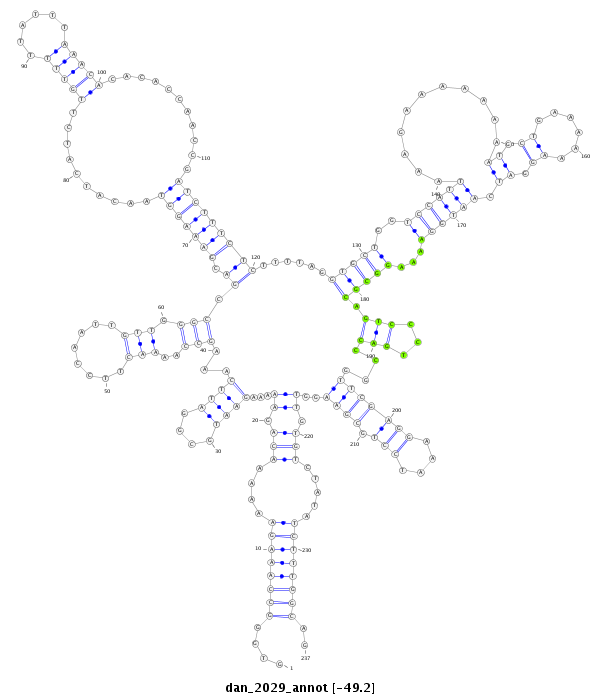
ID:dan_2029 |
Coordinate:scaffold_13266:15416952-15417188 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -49.3 | -49.3 | -49.2 |
 |
 |
 |
exon [dana_GLEANR_13584:1]; CDS [Dana\GF13571-cds]; CDS [Dana\GF13571-cds]; exon [dana_GLEANR_13584:2]; intron [Dana\GF13571-in]
No Repeatable elements found
| ##################################################---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------################################################## ATCAAATCCCTCCAAAAAAAGGCCACTGGACACATCTAAAAATGACTCTGGTGGGCCAAAGAAAAAACAGAAAAAGAATGCGGATTCAAAGCCAAAACTTCCAATTGTTGGGCCGACGAAAGGTAACATCATCTTGTTTTTATTTAAACACACACCAACCGATCTTTCTCTTTTAGGTGCTGGTCCATTAAAGAAAAAAAATGCTGAAAAAAGGATCAATGGAAAAGGCGCAGTCCCTGACCCGGTTCGAGGAAATCCTGCGAAGGTTGTGTCTATATCTTTGGCAGACAAAGGATTCTATGAATCTGATGCAGACAGTCAGGACCAAGAGGCTGTG **************************************************....((((((((....(((.((......((((((((....(((..(((........))).))).((.(((((((..........(((((......)))))...........)))))))))......(((((..((((((...........((.((......)).)).))))))...)))))..(((.(((......))).)))))))).)))....)).))).....))))))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M058 embryo |
V039 embryo |
V055 head |
M044 female body |
V105 male body |
|---|---|---|---|---|---|---|---|---|---|---|
| ............................................................GAAAAAACAGAAAAAGAATCCC............................................................................................................................................................................................................................................................... | 22 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................................AAAAATGACTCTGGTGGGCC........................................................................................................................................................................................................................................................................................ | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................................................AAAAGGCGCAGTCCCTGACCC.............................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................GATGCAGACAGTCAGGACCAAGAGGC.... | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................................................................................................................GATGCAGACAGTCAGGACC........... | 19 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................TTTATTTAAACAGACACCA.................................................................................................................................................................................... | 19 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................AAACTTCCAATTGCTGAGC................................................................................................................................................................................................................................ | 19 | 2 | 5 | 0.20 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................AAGAAAAAGCAGAAAAAGAA................................................................................................................................................................................................................................................................... | 20 | 1 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................................................................................................................................................................................TCGAGGAAGTCCTGAGAAGGG...................................................................... | 21 | 3 | 5 | 0.20 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................AAGAAAGAACAGAAAAAGA.................................................................................................................................................................................................................................................................... | 19 | 1 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................AAGAAAAGACAGAAAAAGA.................................................................................................................................................................................................................................................................... | 19 | 1 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................TGGTTTTATATAAAAACACACC..................................................................................................................................................................................... | 22 | 3 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................TCTAAAGACGCACCAACCGA............................................................................................................................................................................... | 20 | 3 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................................ACAAAGAGAAAACAGAAAAAGA.................................................................................................................................................................................................................................................................... | 22 | 2 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................................................................................................................................CTTCGTAGGTTGTGTCTAC............................................................. | 19 | 3 | 12 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 |
| ........................................................................................................................................................................TCTTGTGGGTGGTGGTCCAT..................................................................................................................................................... | 20 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..................................................................................................................................................................................................................................TGCGCAGTCGCTGACCGGG............................................................................................ | 19 | 3 | 18 | 0.06 | 1 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................................................ACGCAAGGGCACATCATCT........................................................................................................................................................................................................... | 19 | 3 | 19 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| .............................................................AAAAAACAGAAAAAGAGT.................................................................................................................................................................................................................................................................. | 18 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................AAAGAAAAAACAGAAAAAAAA................................................................................................................................................................................................................................................................... | 21 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................AAGAAAAAACAGAAAGAGG.................................................................................................................................................................................................................................................................... | 19 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................AAGAAAAAAGAGAAAAAGA.................................................................................................................................................................................................................................................................... | 19 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............AAAAAAAGCCACGGGACCCA............................................................................................................................................................................................................................................................................................................... | 20 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................ACACCCCGATCGATCTTTC......................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................................AAAGAAAAAACAGAGGAAGAA................................................................................................................................................................................................................................................................... | 21 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| .........................................................AAAGAAAAAACCGAGAAAGA.................................................................................................................................................................................................................................................................... | 20 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................................AAAGAAAGAACATAAAAAGA.................................................................................................................................................................................................................................................................... | 20 | 2 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................................................ATCAATTTTAGGTGCTGGT......................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
|
TAGTTTAGGGAGGTTTTTTTCCGGTGACCTGTGTAGATTTTTACTGAGACCACCCGGTTTCTTTTTTGTCTTTTTCTTACGCCTAAGTTTCGGTTTTGAAGGTTAACAACCCGGCTGCTTTCCATTGTAGTAGAACAAAAATAAATTTGTGTGTGGTTGGCTAGAAAGAGAAAATCCACGACCAGGTAATTTCTTTTTTTTACGACTTTTTTCCTAGTTACCTTTTCCGCGTCAGGGACTGGGCCAAGCTCCTTTAGGACGCTTCCAACACAGATATAGAAACCGTCTGTTTCCTAAGATACTTAGACTACGTCTGTCAGTCCTGGTTCTCCGACAC
**************************************************....((((((((....(((.((......((((((((....(((..(((........))).))).((.(((((((..........(((((......)))))...........)))))))))......(((((..((((((...........((.((......)).)).))))))...)))))..(((.(((......))).)))))))).)))....)).))).....))))))))..************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M044 female body |
V105 male body |
M058 embryo |
V055 head |
V106 head |
|---|---|---|---|---|---|---|---|---|---|---|
| AAGTTTAGGGAGGTTTTTTT............................................................................................................................................................................................................................................................................................................................. | 20 | 1 | 1 | 4.00 | 4 | 2 | 2 | 0 | 0 | 0 |
| AAGTTTAGGGAGGTTTTTT.............................................................................................................................................................................................................................................................................................................................. | 19 | 1 | 1 | 2.00 | 2 | 1 | 1 | 0 | 0 | 0 |
| ...............................................................................................................................................AATTTTTTTGTGGTGGGCTAG............................................................................................................................................................................. | 21 | 3 | 7 | 1.14 | 8 | 8 | 0 | 0 | 0 | 0 |
| AAGTTTAGGGAGGTTTTTTTG............................................................................................................................................................................................................................................................................................................................ | 21 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| AAGTTTAGGGAGGTTTTTTTGC........................................................................................................................................................................................................................................................................................................................... | 22 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............TTTTTTTCCGGCGACCTGT................................................................................................................................................................................................................................................................................................................. | 19 | 1 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ................................................ACCACCCGGTTTCTTTAT............................................................................................................................................................................................................................................................................... | 18 | 1 | 2 | 1.00 | 2 | 0 | 0 | 2 | 0 | 0 |
| .AGTTTAGGGAGGTTTTTTTTG........................................................................................................................................................................................................................................................................................................................... | 21 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| AAGTTTAGGGAGGTTTTT............................................................................................................................................................................................................................................................................................................................... | 18 | 1 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................AAGCCCACGACCAGGGAATTT................................................................................................................................................. | 21 | 3 | 4 | 0.25 | 1 | 0 | 0 | 1 | 0 | 0 |
| AAGTTTAGGGAGGTTTTTTTTG........................................................................................................................................................................................................................................................................................................................... | 22 | 3 | 17 | 0.24 | 4 | 0 | 4 | 0 | 0 | 0 |
| ...............TTTTTCCTGTGACCCGTTTAG............................................................................................................................................................................................................................................................................................................. | 21 | 3 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..GTTTAGGGAGGTTTTTTTTG........................................................................................................................................................................................................................................................................................................................... | 20 | 2 | 9 | 0.11 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................................TCTTACGCTGAAGTTTCGGA................................................................................................................................................................................................................................................... | 20 | 3 | 13 | 0.08 | 1 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................................................................TTGTGGTGGGCTAGAAA.......................................................................................................................................................................... | 17 | 2 | 17 | 0.06 | 1 | 0 | 0 | 0 | 1 | 0 |
| .AGTTTAGGGAGGCTTCTGT............................................................................................................................................................................................................................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................TTTGTGGTGGGCTAGACAG......................................................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| .AGTTTAGGGAGGTTTTTTTTGC.......................................................................................................................................................................................................................................................................................................................... | 22 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 |
| .....................................................................................................................................................................................CCAGGTAATTTTCTTTTTT......................................................................................................................................... | 19 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droAna3 | scaffold_13266:15416902-15417238 + | dan_2029 | ATCAAA------TCCC---------TCCAAAAAAAGGCCACTGGACACATCTAAAAATGACTCTGGTGGGCCAAAGAAAAAACAGAAAAAGAATGCGGATTCAAAGCCAAAACTTCCAATTGTTGGGCCGAC---GAAAGGTAACATCATCTTGTTTTTATTTAAACACACACCAACCGATCTTTCTCTTTTAGGTGCTGGTCCATTAAAGAAAAAAAATGCTGAAAAAAGGATCAATGGAAAAGGCGCAGTCCCTGACCC---------------------------------GGTTCGAGGAAATCCTGCGAAGGTTGTGTCTATATCTTTGGCAGACAAAGGATTCTATGAATCTGATGCAGACAGTCAGGACCAAGAGGCTGTG |
| droBip1 | scf7180000396547:1632091-1632448 - | ATCCCAGCCAACTCCCAAGAACAAGACCAAAAAAAGGCCATTGGACTCATCTAAAGATAAATCTGGTGGTCCAAAGAAAAAGCAGAAAAAG------------------------------GTTGCGCCGACGACTAAAGGTAACTTCATCTTGTTTTTATTTAAGCACTCGCCAATCAAGCTTTTGTTTTTAGGAAAAGCTCCACTAAAAAATAAAAAGTCTGAGAAAAGAATAAATGGAAAAGGCCCAGTTTCCGTCCCAAAAAGTTCCATAAATGGACGCCAAGGAGCGTCGACTCCAGCAAAACCTGGGAAGGTAGTCCAAATCGCTTTTGCAGGCAATGGATTTGGCGGATCTGAGGAAGACGATCAGGACGAAGAAGTTTTG | |
| droEle1 | scf7180000491107:1000527-1000563 - | T----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ATGGATTCGATGAATTCA--GAGGACGATTG---------GGCTGAA |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEle1 |
|
Generated: 05/18/2015 at 03:32 AM