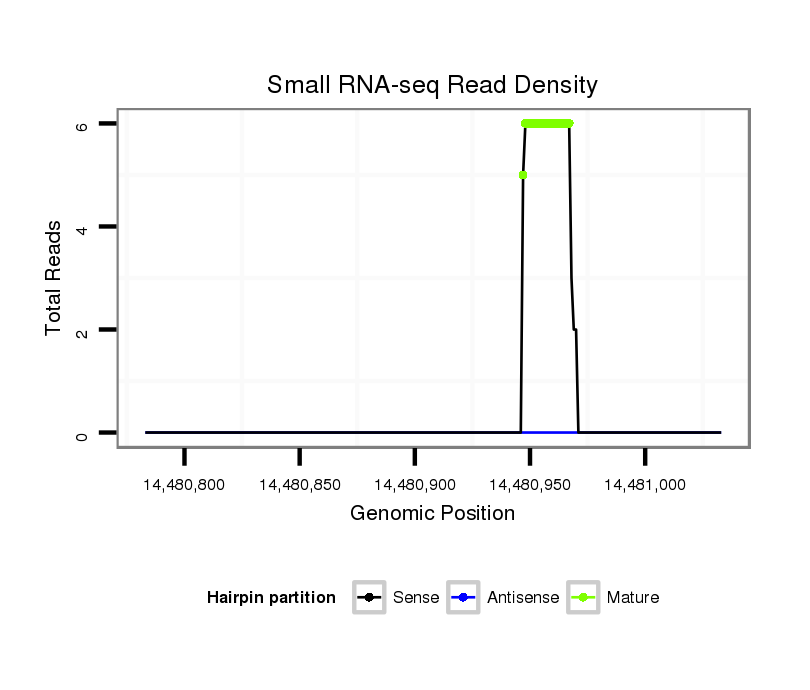
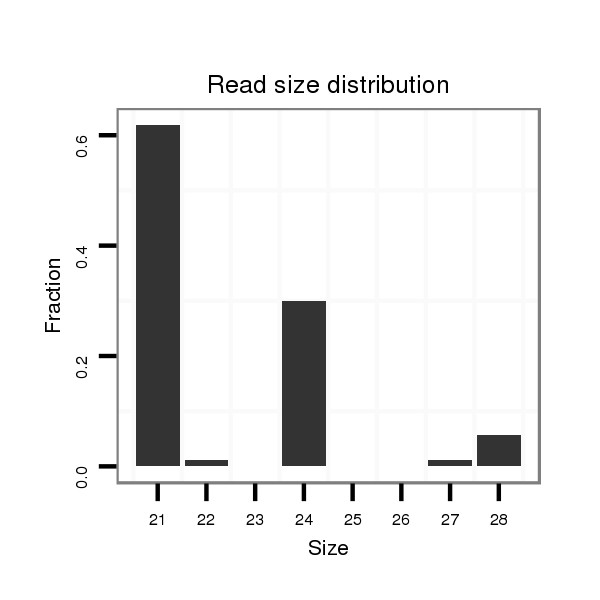
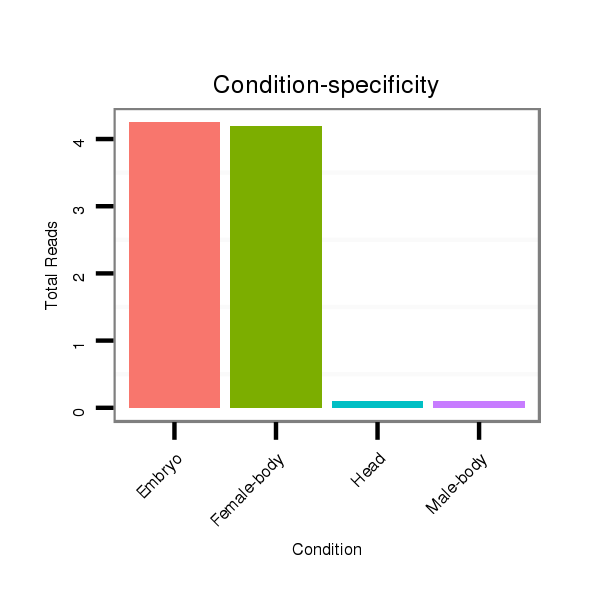
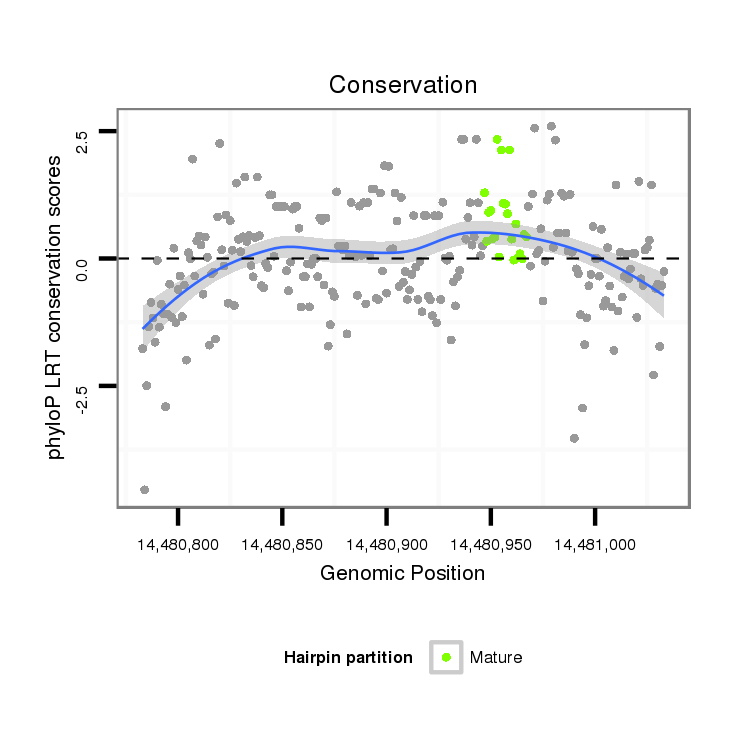
| droAna3 |
scaffold_13266:14480783-14481033 + |
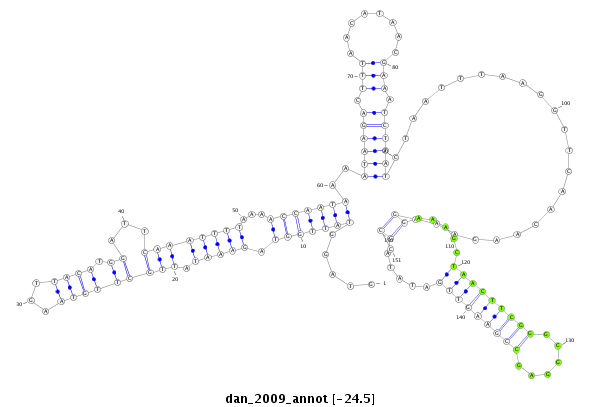
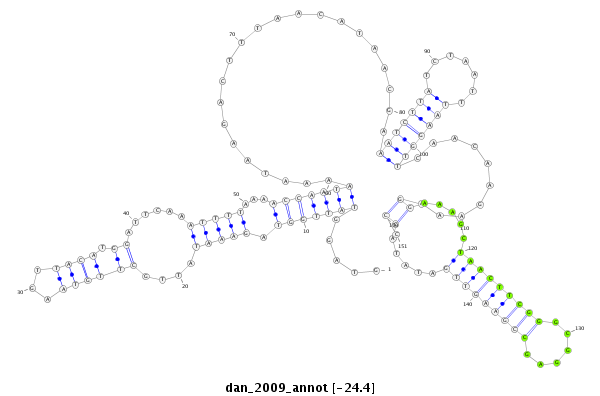
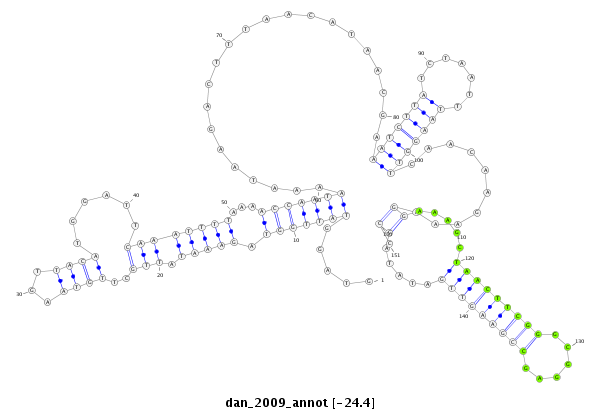
dan_2009 |
TCCAACCAATG-GAAAG---CGGATCTGGCCAGTATTGGGCAGTTTATCCCCAAGTAGGTATTGG-------------------------------TA--GAAATATTGCTTGTAAGTTACA-------------------------------------------------------TGGA---------------------------------------------------TTC------AAATTT---------------------------------------------------------------------------------------------------------------------T-----------AAAACCAATAAAATAA-------------------------------------------------------------------------------------------------GACTTTAACATAACGAAATCTTATCTAATTTAAGGTTCAACAAGAAAGGAAAGCTAACTTCGGGCGGAGCCGAAGTTGATATACCCTTGCAGTTCAGT------CGCAGTCCG----------C---------TAGGTGG-CGC-----------------------CACCCA--TCTTATTTT----ATT |
| droBip1 |
scf7180000396715:16694-16888 + |
|
TCTGTCCAATA-CTCAA---CAGGT--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GATTACTTTTCTCGCCAATTTGGTGAATGTGGTAATCTGAATTGGTTGTGCAGTTTCTCATCCAAAATATA--------A-----------AAAATCAAAAAAAA---------------------------------------------------------------------------------------------------------------TTGAATTATTGATTCAATTAATGTTTAACAAGAAAGCAAAGCTAACTTCGGGCGGAGCCGAAGTTGATATACCCTTGCAGCTAAAA------CC------------------------------------------------------------------------------------- |
| droKik1 |
scf7180000301628:7744-7985 + |
|
TGAAA----------AG---C-----AGGCTGGTATTAACCAAATCAGCCAACACAAGGTGGCA-CCGTGAGA--------------AAATATCTT---------------------------------------------------------------------------------------------------------------------------------TGAACTAGTTTCAATTG-----ATAATTTTAAAATAATG---------------------------------------------------------------------------------------------------GG--CA-ATAAAA-------------------------------------------------TA------TTGAAATTA--------TTAAGAGTAGTA----------------GTAGTAGACCAAAAT--------------------------AAACAATAAACAAGAAAAG-AAGCTAACTTCGGC--ACGCCGAAGTTTGTATACCCTTGCAAATATTA------TT----TCA----------T---------TAAATT-----------------------T----TACCTA-TACTTATTAT----GTT |
| droFic1 |
scf7180000449015:848-1139 + |
|
TTTAAGCGGAA-AGAAG---AGGATTTA-------------------------------------------------------------------------------------AGGACA--GTATATGGGAGCCAGTTCTTGGGATACATTATT-----GATTCTAATGGTAGAATATGGAGTTAAATTGGTTAATAATTACAGAGTAGGGCTTTGGGAACTTCA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGAAAAGAT-----------------------------------------------------TTCATAGAACAAAAT----------------------------A-TATAAACAAGAAAGG-AAGCTAGCTTCGGC--AAGCCGAAGCTTATATACCCTTGCAGTCATCAAATATGTC-------G-----AACAAAACGCATAAAGGCTAGACAACATCGAAGCTAAGACGATTTTTCTATTCATTTTCCATATT----TTT |
| droEle1 |
scf7180000490370:2563-2770 - |
|
TTTACCCTGAT-CAAGG---TATGCG----------------------------------------------------------------------TAAATAAATAAGACTTGCAAAAA--A-------------------------------------------------------TGTA---------------------------------------------------TGT------GAATTT-------------------------------T-------------------------------------------------------------------------ATAAGCTACTGCG--CG-------------------------------------------------------------AAA------TATAAATAACATTTTGTGTAA--T-------------------------------------------------------TATA-TGAAAACAAGAAAGG--AGCAAACTTCGGC--AAGCCGAAGTTTATATACCCTTGCAGCTATTG------CA--------AGAATTAAAT---------ATTTTTGAAGACATTA-----------------------A--AATTATGATCT-ACTT |
| droRho1 |
scf7180000762630:45131-45349 - |
|
CA------ATG-GAAAGACAACGTTTTGGACAGTTTCAAGCAGATAGC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TTTAAAACTGAGGGACTAGTTTGCGTT-------------------------------------------------------------------------GCAAACTTTTGAC--TG-------------------------------------------------------------AAA------TTATAATAACC---TCTGCAA--GGGTATCAAAAT---------AGAGAA---------AT--------------------AGCTAAAAACAAGAAAGGAAAGCTAACTTCGGGCGGAGCCGAAGTTGATATACCCTTGCAGAATACT------CT------G-CAAATTATAT-------------------A-----------------------CTCCAA--AATT--------ATTC |
| droBia1 |
scf7180000301527:8251-8471 + |
|
CCCAACGAACG-ACAAG---AGGGACGCGCAGCCAGAATGCAGTCT----------------------------------------------------------------------------------------------TAGTATAC--TGCTAATCAAATTTTAATGT-------------------------------------------------------TAAGATTCGT------AAACTT-------------------------------ATTATTAGTC----------------------------------------------------------TCAAAATAAA--------GAGAAGATTTAA-------------------------------------------------AATTAA--AAGCAA------AG-------------------------------TTAATA-----------------------------------AAACAAAAAACAAGAAAGG-AAGTTAACTTCGTC--AATCCAAACTTTGTATACCCTTGTAGTTATAA------GC-------AATAAT-----------------------------------------------------------C---AACTTTAGT |
| droTak1 |
scf7180000414479:56857-57101 - |
|
AGAGAAA----------------ATATGGTCAGACTGGGGCA-----------------------CGGATAGGGATTATTAATATTAAAATTTTTT---------------------------------------------------------------------------------------------------------------------------------TTATGTATACACACCTG-----ATCATATTAAAAATGAGG----------------------------------------------------------------------------------------TAAATTCTTGTC--AA-------------------------------------------------------------AAA------CCAAAG-----------------TTACATGCGGTTTTAA-------------------------------------------TTAAAAAACAAGAAAGG-AAGCTAGCGTCGGC--CAGCCGAAGCTTATATACCCTTGCAGATAATT------CCTA----T-TAATTTACAAATCGCAAAAATGGTA---------------------ATT----TTCCTA--TTATTTCTC----ATT |
| droEug1 |
scf7180000409111:41118-41191 - |
|
AATAATAT-----TTTA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------A-------------------------------------------CAACAAAATAATTAAAG-AAGCTAACTTCGGC--AAGCCAAAGTTTATATACCCCTGCAGCTTA----------------------------------------------------------------------------------------------- |
| dm3 |
chr2R:10132035-10132091 + |
|
TCCAACCAATG-GAAGG---CGGATCTGGTCAGCATCGGGCAGTTTATTCCAAAGTGGGTA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droSim2 |
2r:10835001-10835057 + |
|
TCCAACCAATG-GAAGG---CGGATCTGGTCAGCATCGGACAGTTTATTCCAAAGTGGGTA------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| droSec2 |
scaffold_1:5681320-5681408 - |
|
GGCTTTTTCA----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ACACCAGATTT------TGGCA---------------------------------------------AGAACAAA---------------------------------GCTAACAGGAAGGG-AAATTAACTGCGAC--AAGCCGAAGTTTGTATGCCCTTGCAGT-------------------------------------------------------------------------------------------------- |
| droYak3 |
2R:10068961-10069020 + |
|
TCCAACCAATG-GAAGG---CGGATCTGGTCAGCATTGGACAGTTTATTCCAAAGTGTGTA-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TT-----------------------------------------------------------------------------------------------------------G |
| droEre2 |
scaffold_4845:15711597-15711749 - |
|
TCCAACCAATG-GAAGG---CGGATCTGGTCAGCATTGGACAGTTCATCCCAAAGTGTGTA--------------------------------------------------------------------------------------------------------------------------------------------------------------------TTACATAGAC------AAATTT-------------------------------TTTAATCGGC----------------------------------------------------------TTCACAT-------------------------------------------------------------------------------------------------------------ATACATCCGATTTGAA---------TG---------AA-------------------------------------------------------------TACTACATATACTTTTATAAACATAA------AC-------AA------------------------------------------------------------TCTT---ATCTTATCT |
| dp5 |
3:17382729-17382786 + |
|
TCCAACCAGTG-GAAGG---CAGACCTGGCCAGCATTGGCCAGTTCATACCCAAGTGAGTA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------T----------------------------------------------------------------------------------------------- |
| droPer2 |
scaffold_3:5054154-5054228 - |
|
AAGGTCCATGA-AGTTG---AAAATTT-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------A-----------AAAGTCAATAAAATCA-------------------------------------------------------------------------------------------------GATTTAA-------------------------------------------------------------ATATAATTTGATTTACTTTGATGTCTA-----------------------------------------------------------------------------------------------A |
| droWil2 |
scf2_1100000004510:2021075-2021242 - |
|
ATTCAGAAACTTAACCA---CAAGCCAAGTCATTAACTAAAATGTTAT-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------CAGCATTTAATTTAAATTACA--------GAGAA------------------AACGA---TACATTTAACTATATATTGAT-------------------------------------------------------------------AAACTAGATTTAA-------------------------ACATAAAAT-ACCAAAG-AAGCTAACTTCGGC-TATGCCGAAGTTTATATACCCTTGCAGT-------------------------------------------------------------------------------------------------- |
| droVir3 |
scaffold_12941:221526-221689 + |
|
AAGTCTGAGTG-AGAAG---AGTGCGT-----GTAATGGGCA--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AGTGC---------------------------------------------------------------------------------------------------------------------G-----------GAGGCCGAAAGAACAAGAT----------------------------------------------------------------------------------------------CACTAAAAGAATACCACACATTAAGTGATTTTATATACAACGAGAAAGAAGGGCTATCTTCGGC--ACGCCAAAGATCTAATACCCTTGCAGACCCAA------CC-----------------------------------------------------------------------TT--TTC----ATA |
| droMoj3 |
scaffold_6496:11196095-11196164 - |
|
TCTAATCAATG-GAAGG---CGGATCTGGCCAGCATCGGACAGTTCATACCCAAGTAAATATAGA-------------------------------TA--T-----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------AAA----TAT |
| droGri2 |
scaffold_15110:1256437-1256648 - |
|
ATTCTCAAGTA-TATAG---T---------CA----------------------------------------------------------------TATCTAAATATAGCTT-----------------------------------------------------------------------------------------------------------------TTACGTGCATGCAACTA------------------------------------AATAGGTGTA----------------------------------------------------------TATAAAAAAA--------GTAAAAACAAAA-------------------------------------------------AA------TAGCAA------TA---ATAAGAG-----T-----------------------------T----------------------------A-TATAAACAAGAAAGAACGGCGATCTTAGGT--AAGACGAAGACGAAATGCCCTTGCAGACAATG------TC-------GATAGTTAAAC---------TACATGAAAAACCCAA-----------------------A--ACTTATGATATTTTGT |