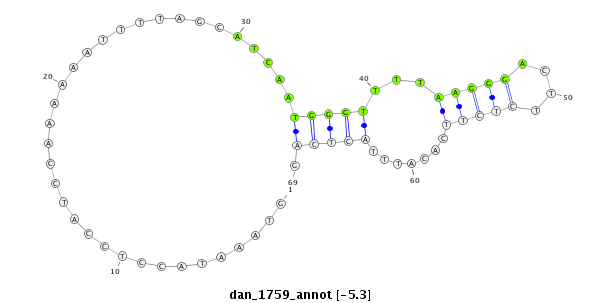
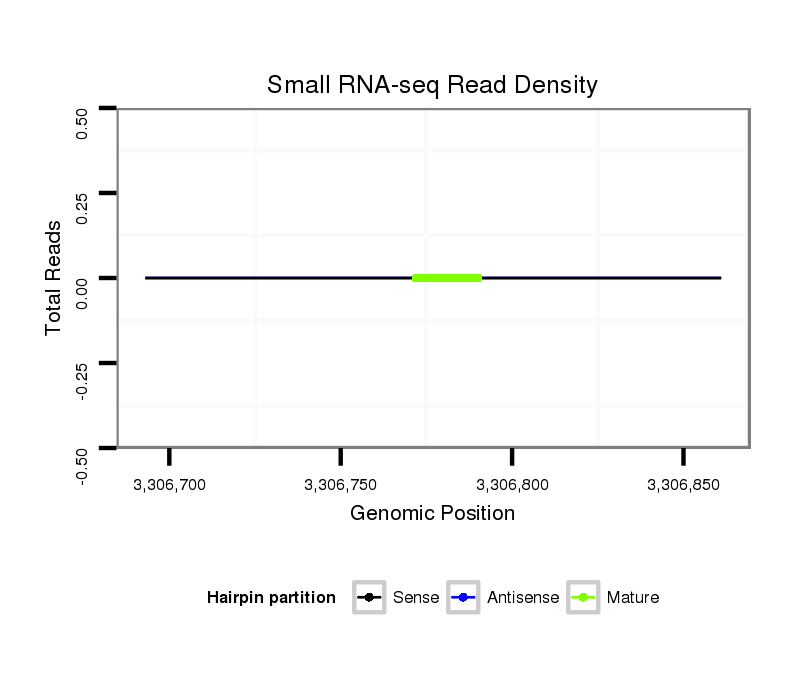
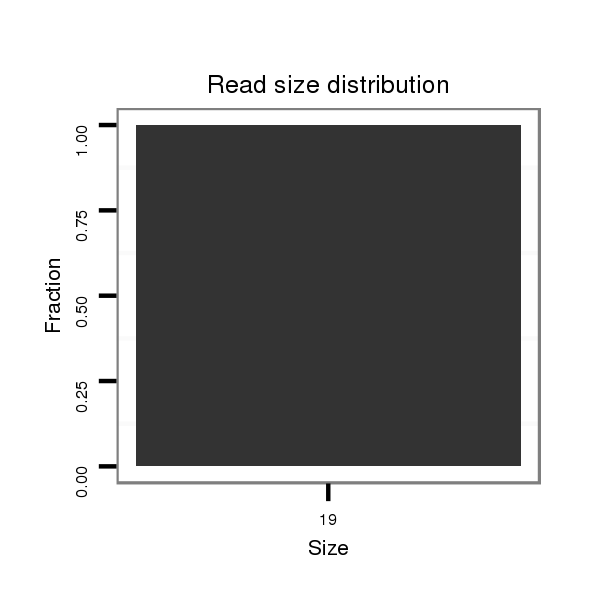
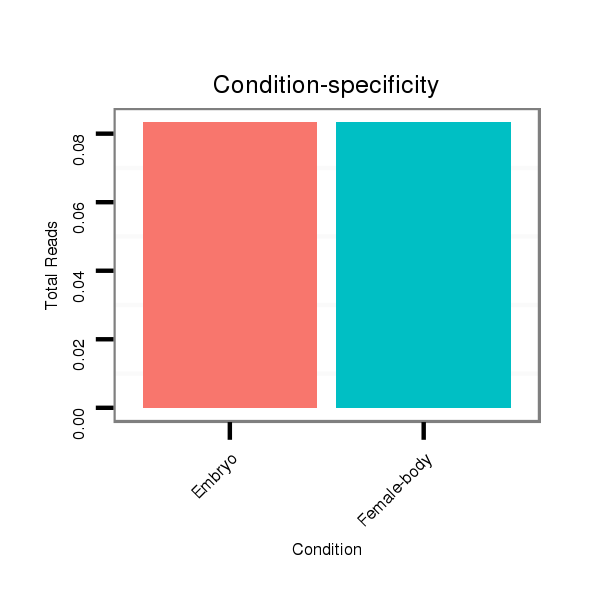
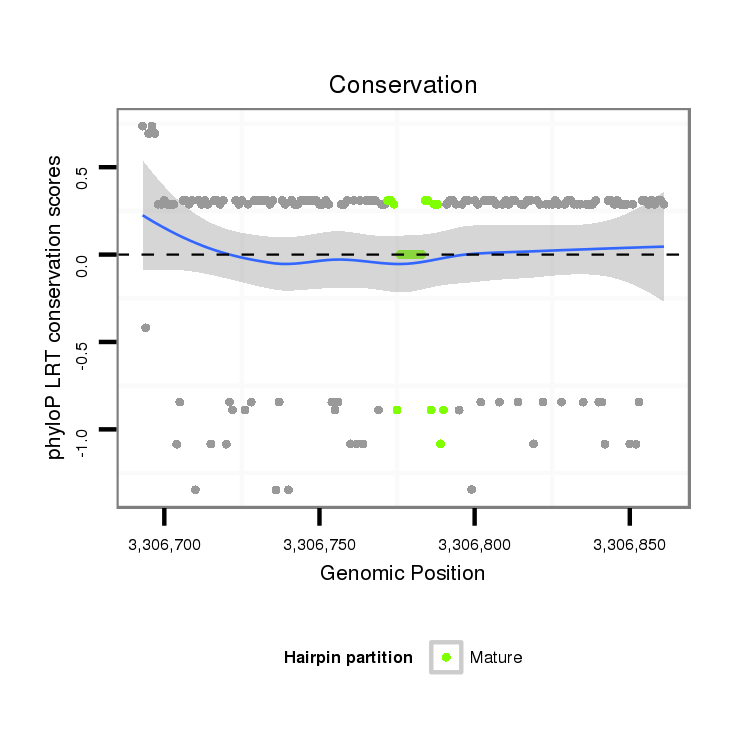
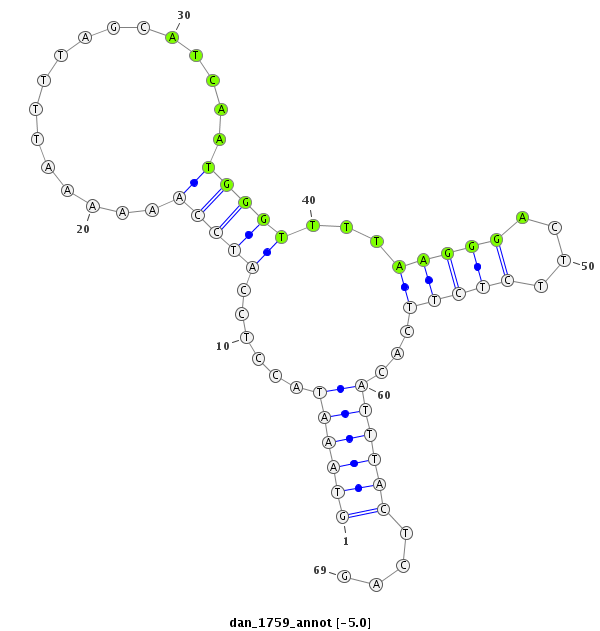
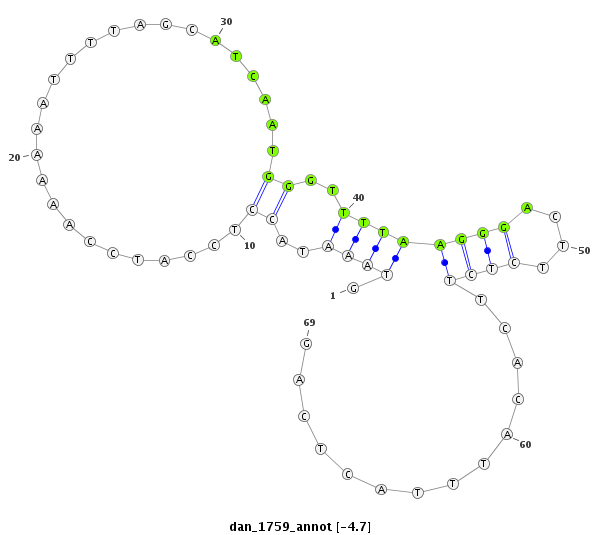
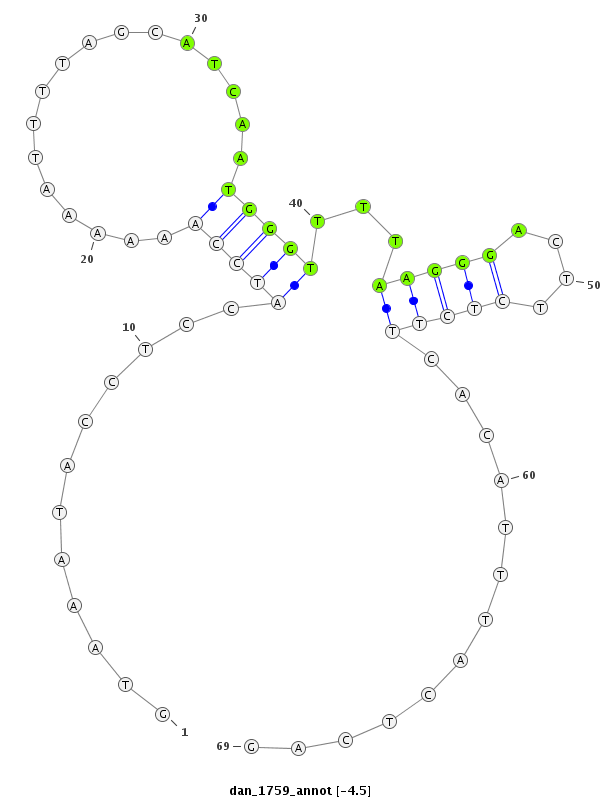
ID:dan_1759 |
Coordinate:scaffold_13266:3306743-3306811 + |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -5.0 | -4.7 | -4.5 |
 |
 |
 |
CDS [Dana\GF12700-cds]; CDS [Dana\GF12700-cds]; exon [dana_GLEANR_12718:1]; exon [dana_GLEANR_12718:2]; intron [Dana\GF12700-in]
No Repeatable elements found
| ##################################################---------------------------------------------------------------------################################################## ACCTGGCAGCGCTATGAGAGAGAAAGATGAAGTACGTTTTTCAGATCGTGGTAAATACCTCCATCCAAAAAATTTTAGCATCAATGGGTTTTAAGGGACTTCTCTTCACATTTACTCAGATGCCATATGCGGCTGCGTTCCCGGCCTGTCAAGTCCCAGCCAAGTCTTC **************************************************..................................(((((...(((((....))))).......))))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M058 embryo |
V055 head |
V039 embryo |
M044 female body |
|---|---|---|---|---|---|---|---|---|---|
| ..................................................GTAAAGACCCCCACCCAAAAAA................................................................................................. | 22 | 3 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 |
| ...........ATATGCGAGAGAAAGATGAAGGA....................................................................................................................................... | 23 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 |
| ...............................................................................ATCAATGGGTTTTAAGGGA....................................................................... | 19 | 0 | 12 | 0.17 | 2 | 1 | 0 | 0 | 1 |
| ................................................................CTAAAAAATTTTAGCACCA...................................................................................... | 19 | 2 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 |
| ...........................................................................TTGCATCAATGGGTTTTAAGGGAC...................................................................... | 24 | 1 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 |
| ......................................TATCAGATCGAGGTAAAT................................................................................................................. | 18 | 2 | 10 | 0.10 | 1 | 0 | 0 | 1 | 0 |
| ....................................................AAATACCTACACCCAAAAAA................................................................................................. | 20 | 2 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 |
| ....................................TGTTTGAGCTCGTGGTAAA.................................................................................................................. | 19 | 3 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 |
| .......AGGGCTATGAGTGACAAAG............................................................................................................................................... | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 |
| ....................................TATTTGAGATCGTGGTAA................................................................................................................... | 18 | 2 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 |
|
TGGACCGTCGCGATACTCTCTCTTTCTACTTCATGCAAAAAGTCTAGCACCATTTATGGAGGTAGGTTTTTTAAAATCGTAGTTACCCAAAATTCCCTGAAGAGAAGTGTAAATGAGTCTACGGTATACGCCGACGCAAGGGCCGGACAGTTCAGGGTCGGTTCAGAAG
**************************************************..................................(((((...(((((....))))).......))))).************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V055 head |
M058 embryo |
M044 female body |
|---|---|---|---|---|---|---|---|---|
| .................CTCTCTTACTACTTCATGTTA................................................................................................................................... | 21 | 3 | 7 | 0.14 | 1 | 1 | 0 | 0 |
| .............................................................................CGTAGTTACCCAAAATTCC......................................................................... | 19 | 0 | 12 | 0.08 | 1 | 0 | 1 | 0 |
| ..............................................................................................CCCTGACGAGAAGTGGAAC........................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 1 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droAna3 | scaffold_13266:3306693-3306861 + | dan_1759 | ACCTGGCAGCGCTATGAGAGAGAAAGATGAAGTACGTTTTTCAGATCGTGGTAAATACCTCCATCCAAAA--------AATTTTAGCATCAATGGGTTTTAAG---------GGACTTCTCTTCACATTTACTCAGATGCCATATGCGGCTGCGTTCCCGGCCTGTC--------AAGTCCCAGCCAAG-TCTTC |
| droBip1 | scf7180000396427:1774713-1774899 + | ACCTGGCAGCGACATGACAGAGCAAGAGATAGTTCATTTTTCACGTCCTGGTAAATACCTCTTCCCACACTGTTGGTGACTTTTTGCATCT--------TATGGTGAGGTTAGTTCTTCACTTGACGTTTACCCAGATACCATCTGTGGCTGTGTTCCCAGCCTACACGAGATGAAAGTCCCCGATAAGATCTTC | |
| droBia1 | scf7180000302292:3062518-3062522 - | AGCTG---------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
||||||||||||||||||||||
| droBip1 |
|
||||||||||||||||||||||
| droBia1 |
|
Generated: 05/18/2015 at 02:57 AM