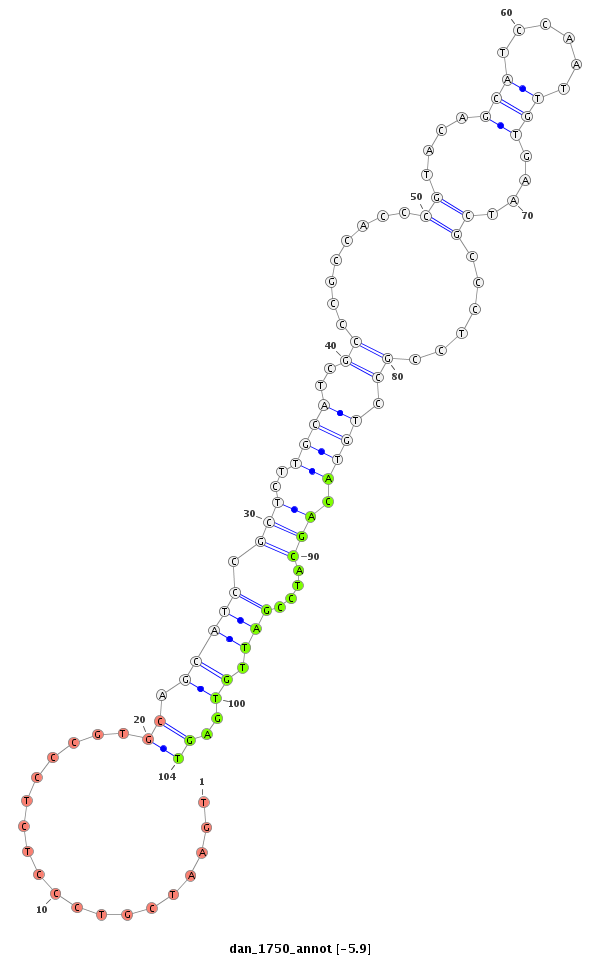
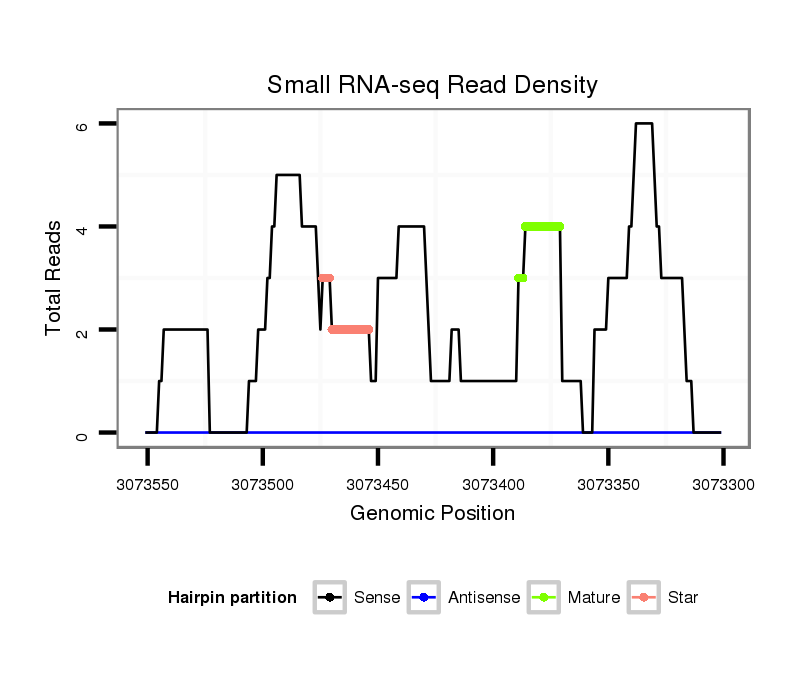
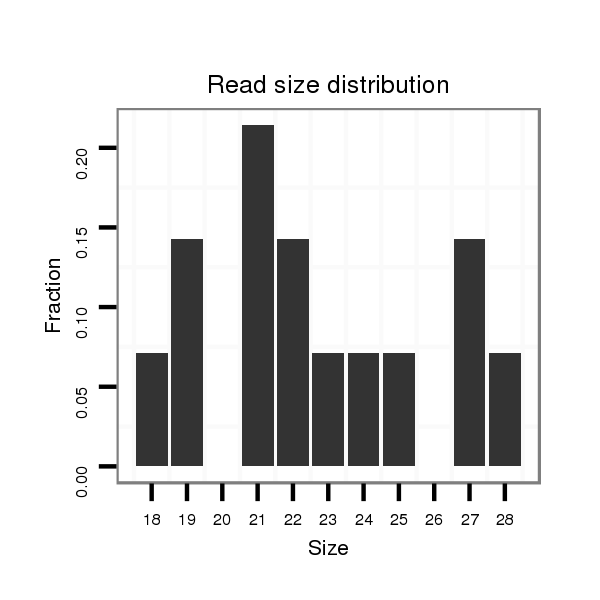

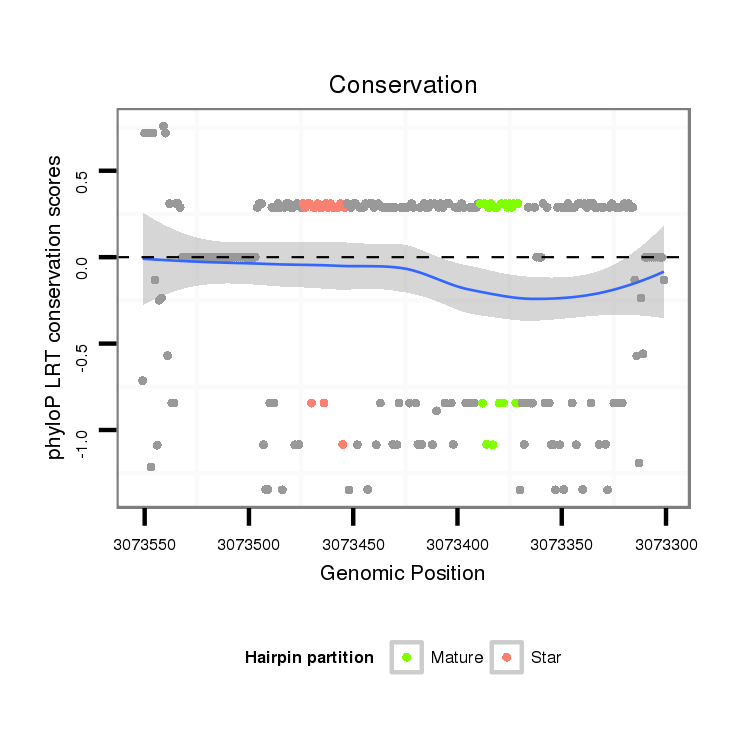
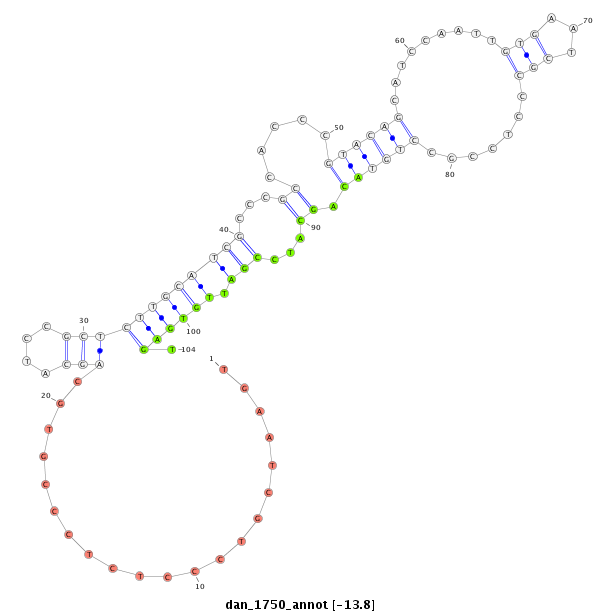
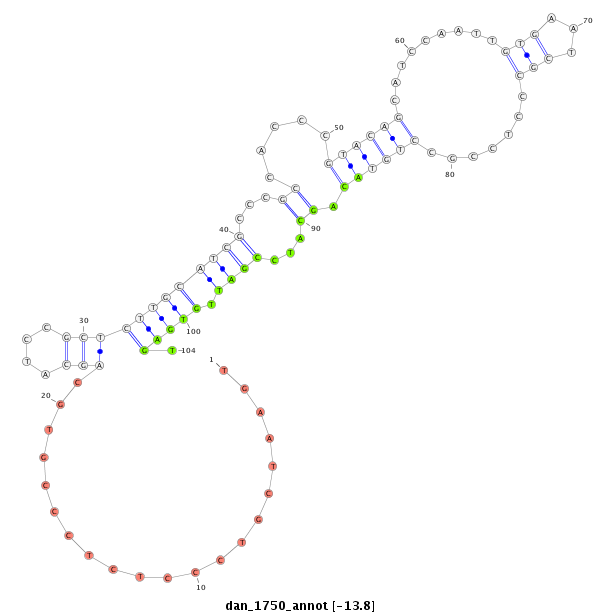
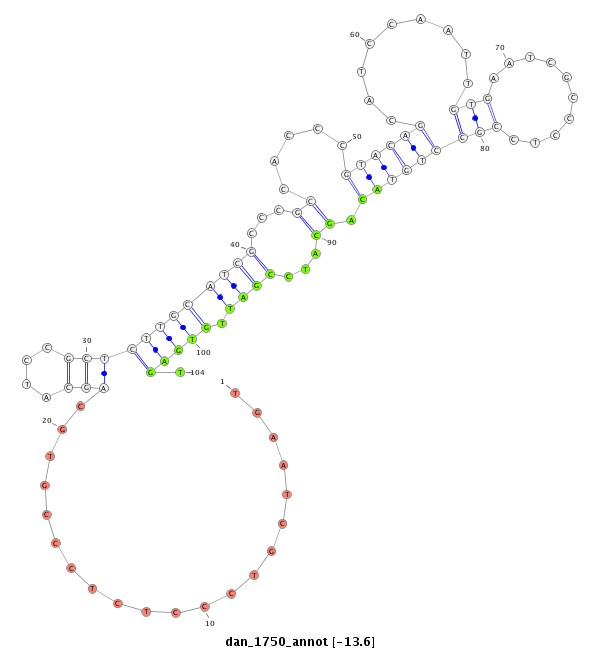
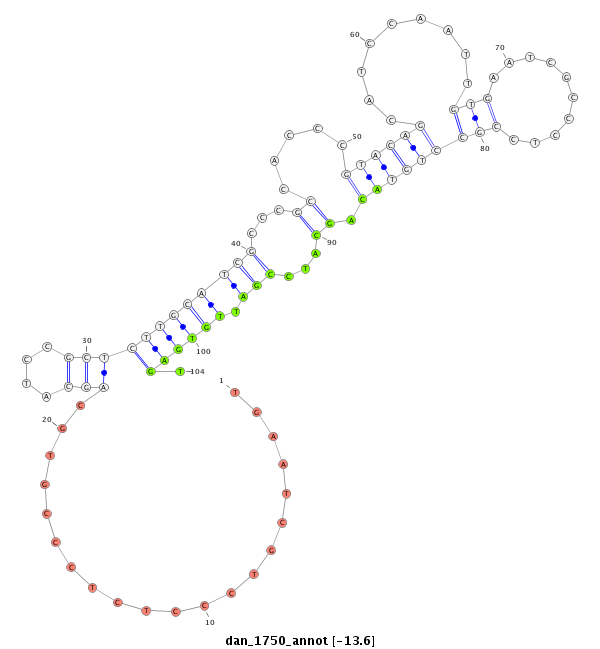
ID:dan_1750 |
Coordinate:scaffold_13266:3073351-3073501 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -13.8 | -13.8 | -13.6 | -13.6 |
 |
 |
 |
 |
exon [dana_GLEANR_12267:3]; CDS [Dana\GF12261-cds]; intron [Dana\GF12261-in]
No Repeatable elements found
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GGGCTCGTCTTCAAGTTACGGTCACAGTAACCAGCACTCGTATGTGTCTCGTTCACAACCCTCCCTGGCATCGCTGGTGAATCGTCCCTCTCCCGTGCAGCATCCGCTCTTGCATCGCCCGCCACCCGTACAGCATCCAATTGTGAATCGCCCTCCGCCTGTACAGCATCCGATTGTGAGTGAGTCCTCACCACCACCGCCGCCTCCGCTTCTGAATCGACCGGCATCCGTACAGGCTCAGCTTCAACAGG *****************************************************************************...................((.(((((.(((..((((..((........((....(((......)))....))......)).)))).)))....))).))..))********************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | M044 female body |
M058 embryo |
V105 male body |
V106 head |
V055 head |
|---|---|---|---|---|---|---|---|---|---|---|
| ..................................................................................................................................................................ACAGCATCCGATTGTGAGT...................................................................... | 19 | 0 | 1 | 2.00 | 2 | 0 | 0 | 2 | 0 | 0 |
| ........CTTCAAGTTACGGTCACAGT............................................................................................................................................................................................................................... | 20 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................................................TGAATCGTCCCTCTCCCGTGC......................................................................................................................................................... | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................................................TGAATCGACCGGCATCCGTACA................. | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................ATCCGCTCTTGCATCGCCCGC................................................................................................................................. | 21 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................ATCCGCTCTTGCATCGCCCGCC................................................................................................................................ | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................TGCATCGCCCGCCACCCGTACAGCATC.................................................................................................................. | 27 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| .....................................................CACAACCCTCCCTGGCATCGCTGGTGAA.......................................................................................................................................................................... | 28 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................GCCTCCGCTTCTGAATCGACC............................. | 21 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................ATCCGCTCTTGCATCGCCCGCCA............................................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................GCTCACAACCCTCCCTGGCAT.................................................................................................................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................................GTCTCGTTCACAACCCTCCCTGG....................................................................................................................................................................................... | 23 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................ACCGCCGCCTCCGCTTCTGAATCGAC.............................. | 26 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................CATCCAATTGTGAATCGCCCTCCGCCT........................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................TCTGAATCGACCGGCATCCGTACAGGCT............. | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................................CTCTGCTTCTGAATCGACCGGCATCC...................... | 26 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................................GTACAGCATCCGATTGTGAGT...................................................................... | 21 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| .....................................................................................................................................................................GCATCCGATTGTGAGTGAGTCCTCA............................................................. | 25 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| ......GTCTTCAAGTTACGGTCACAGT............................................................................................................................................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 0 | 0 | 1 | 0 | 0 |
| .................................................CGTTCACAACCCTCCCTGGCATCGCTG............................................................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 0 | 0 | 0 | 1 | 0 |
| .........................................................ACCCTCCCTGGCATCGCT................................................................................................................................................................................ | 18 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................AACCAGCACTCGTATGTGTCC.......................................................................................................................................................................................................... | 21 | 1 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| .....................................................................................................................................................................................................................GAATCGACCGGCATCCGTACAG................ | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............................................................................AATCGTCCCTCTCCCGTGCAGC...................................................................................................................................................... | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................ACCGCCGCCTCCGCTTCTGAATCGACCGG........................... | 29 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................................CAACCCTCCCTGGCATCGCTGGTG............................................................................................................................................................................ | 24 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................................................TGGCATCGCTGGTCACTCGT...................................................................................................................................................................... | 20 | 2 | 2 | 0.50 | 1 | 0 | 0 | 0 | 0 | 1 |
| .................................................................................................................................................................................GAATGAATCCTCACCACCACC..................................................... | 21 | 2 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................................................TTGTGCGTGAGGCATCACCA.......................................................... | 20 | 3 | 3 | 0.33 | 1 | 0 | 0 | 0 | 0 | 1 |
| ..................................................................................................................................................................................AATGTGTCCTCACCACCA....................................................... | 18 | 2 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................................................CTGGCATCGTTGGGGAAT......................................................................................................................................................................... | 18 | 2 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................TTACAACCGTCGCTGGCATC................................................................................................................................................................................... | 20 | 3 | 7 | 0.14 | 1 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................TCCGCGCTAGCATCACCCG.................................................................................................................................. | 19 | 3 | 10 | 0.10 | 1 | 0 | 0 | 0 | 0 | 1 |
| ...............................................................................................................................................................................................CGCCACCGCCGCCTCCGCT......................................... | 19 | 1 | 14 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................GTGAGGGGTCCCTCTCCCG............................................................................................................................................................ | 19 | 3 | 15 | 0.07 | 1 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................CCTGTCCCGTGGAGCATTC.................................................................................................................................................. | 19 | 3 | 19 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| ............................................................................................................................................................ACCTGTTCAGCATCCGGTT............................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| ................................................................................................................................................GAATCGCCCCCGGCCTGCA........................................................................................ | 19 | 3 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 1 |
| ....................................................................................................................................................................................................................................CGTACAGGTTCAACTTCGA.... | 19 | 3 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
|
CCCGAGCAGAAGTTCAATGCCAGTGTCATTGGTCGTGAGCATACACAGAGCAAGTGTTGGGAGGGACCGTAGCGACCACTTAGCAGGGAGAGGGCACGTCGTAGGCGAGAACGTAGCGGGCGGTGGGCATGTCGTAGGTTAACACTTAGCGGGAGGCGGACATGTCGTAGGCTAACACTCACTCAGGAGTGGTGGTGGCGGCGGAGGCGAAGACTTAGCTGGCCGTAGGCATGTCCGAGTCGAAGTTGTCC
**********************************************************************...................((.(((((.(((..((((..((........((....(((......)))....))......)).)))).)))....))).))..))***************************************************************************** |
Read size | # Mismatch | Hit Count | Total Norm | Total | V055 head |
V105 male body |
M044 female body |
V106 head |
M058 embryo |
|---|---|---|---|---|---|---|---|---|---|---|
| .............................................................................................................................................................................................................GGCGAAGCCTTAGCTGGCCA.......................... | 20 | 2 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................................................................................ACTCAGGGGTGGTGGTTGC.................................................... | 19 | 2 | 3 | 0.67 | 2 | 0 | 0 | 2 | 0 | 0 |
| ..................................................................................GCAGGGAGAGCCCAAGTCGTA.................................................................................................................................................... | 21 | 3 | 3 | 0.67 | 2 | 0 | 0 | 0 | 2 | 0 |
| ....................................................................................................................CTGGCGGTGGGTATGTCCTA................................................................................................................... | 20 | 3 | 5 | 0.60 | 3 | 0 | 3 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................TGGCGTCGGAGGCGAGGACT.................................... | 20 | 2 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................AGTGACATTGATCGTGAGCAG................................................................................................................................................................................................................. | 21 | 3 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................GCTGGCGGTGGGTATGTC...................................................................................................................... | 18 | 2 | 9 | 0.11 | 1 | 0 | 0 | 1 | 0 | 0 |
| ....................................................................................................................................................................................ACTCAGGGGTGGTGGTTG..................................................... | 18 | 2 | 9 | 0.11 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..........TGTTCAATCCCAGTGTCAAT............................................................................................................................................................................................................................. | 20 | 3 | 11 | 0.09 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...............................................GCTCGAGTGTTGGGAGGGA......................................................................................................................................................................................... | 19 | 3 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................................................................CTGGCGGTGGGTATGTC...................................................................................................................... | 17 | 2 | 16 | 0.06 | 1 | 1 | 0 | 0 | 0 | 0 |
| .............................................CGGGGCAAGGGTTGGGAGGG.......................................................................................................................................................................................... | 20 | 3 | 16 | 0.06 | 1 | 0 | 0 | 0 | 0 | 1 |
| ...............................................................................................................................................................................................GTGGTAGCGGCGGAGGCAA......................................... | 19 | 2 | 18 | 0.06 | 1 | 0 | 0 | 1 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droAna3 | scaffold_13266:3073301-3073551 - | dan_1750 | GGGCTCGTCTTCAAGTTACGGTCACAGTAACCAGCACTCGTATGTGTCTCGTTCACAACCCTCCCTGGCATCGCTGGTGAATCGTCCCTCTCCCGTGCAGCATCCGCTCTTGCATCGCCCGCCACCCGTACAGCATCCAATTGTGAATCGCCCTCCGCCTGTACAGCATCCGATTGTGAGTGAGTCCTCACCACCACCGCCGCCTCCGCTTCTGAATCGACCGGCATCCGTACAGGCTCAGCTTCAACAGG |
| droBip1 | scf7180000396427:1538432-1538634 - | AGGCCCGACTTCGAACTAC------------------------------------CAAAGGCCTCTGCCATCGATTGTGAACCGTCCTTCTCCCGTTCACCATACGCTGTTGAACCGCCCTCAGCCCGCACGTCCTCCACTAGTGGATTTCCCTCTACCCGTATATCAGCCAACTGTGAATCGTCCTCC---ATCGAACCAGGCTCTGATTGTGAGTCGCCCTCCACCTGCACAGGCCGAA---------G | |
| droEre2 | scaffold_4929:13464973-13464991 - | AGGCGCAGAGTCG-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------TCGCG---------A |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEre2 |
|
Generated: 05/18/2015 at 02:52 AM