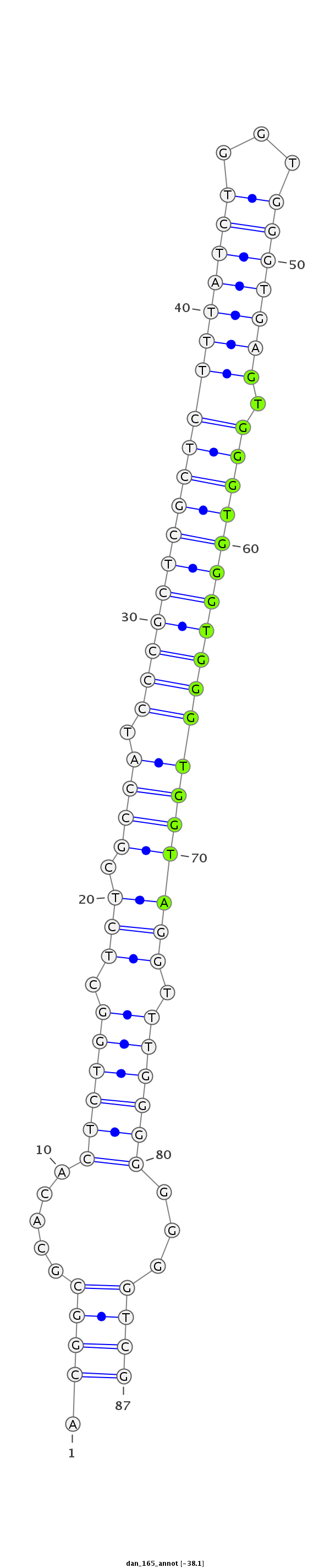
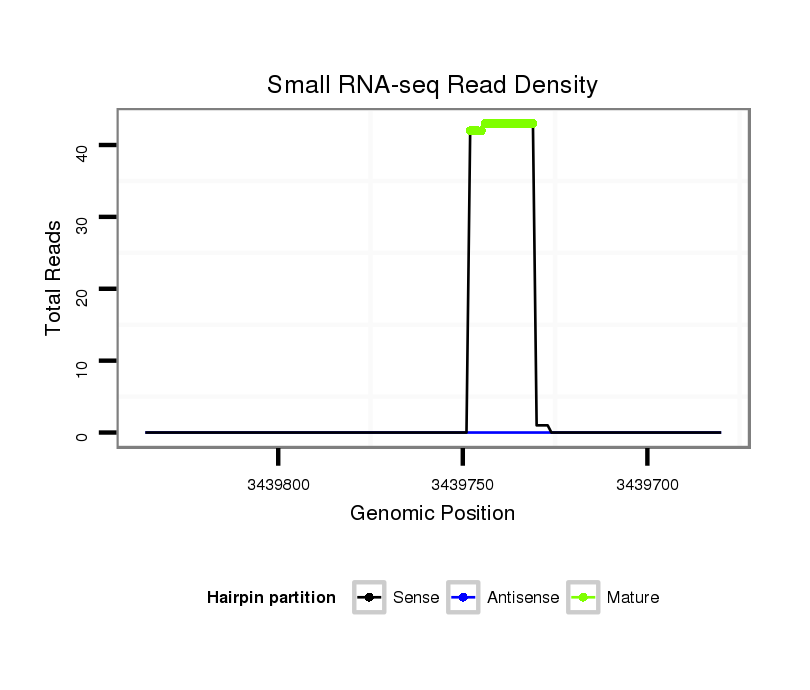
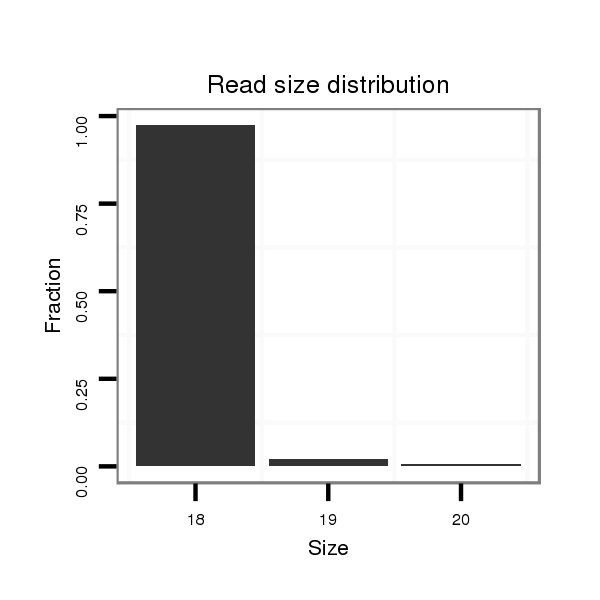
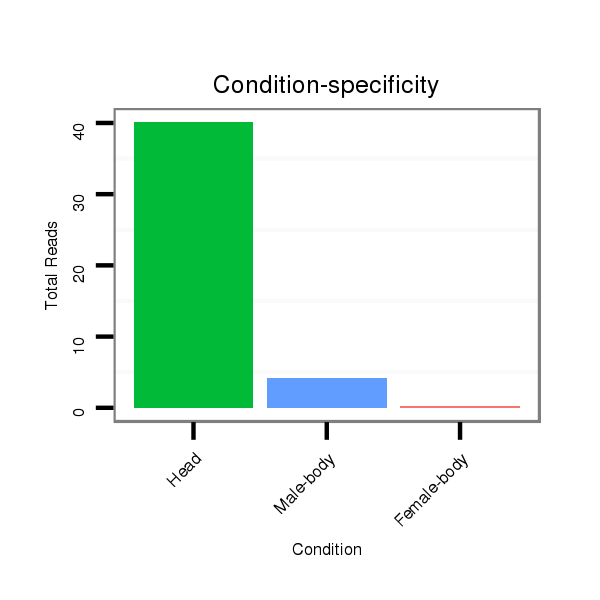
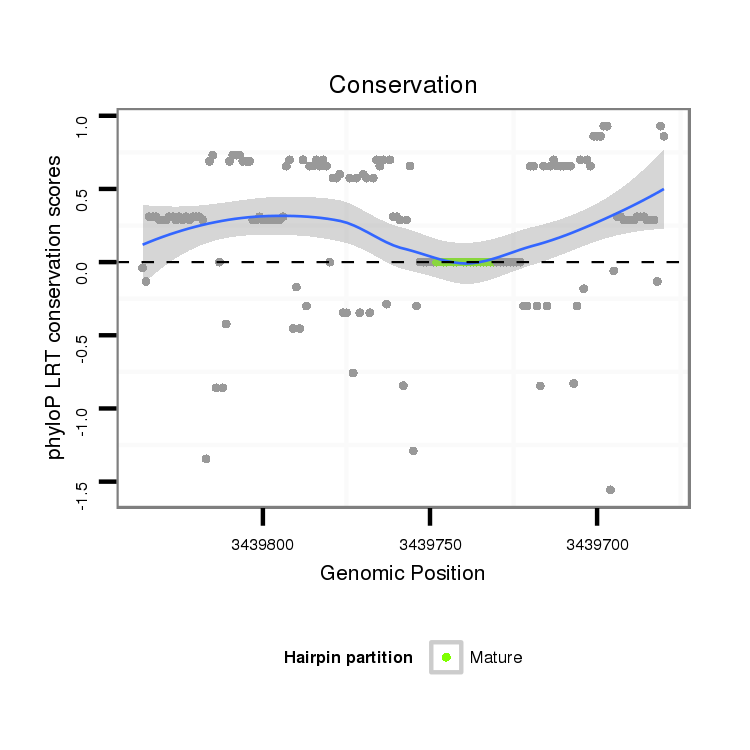
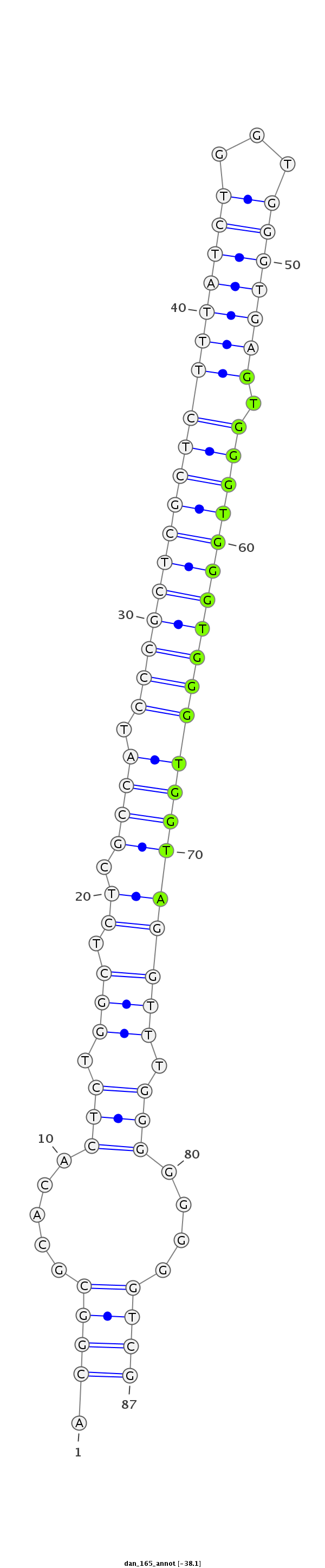
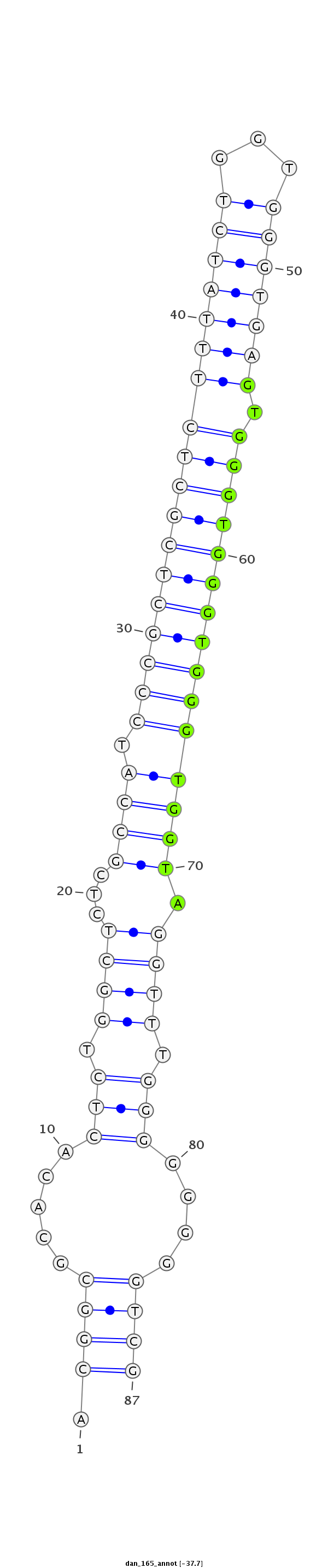
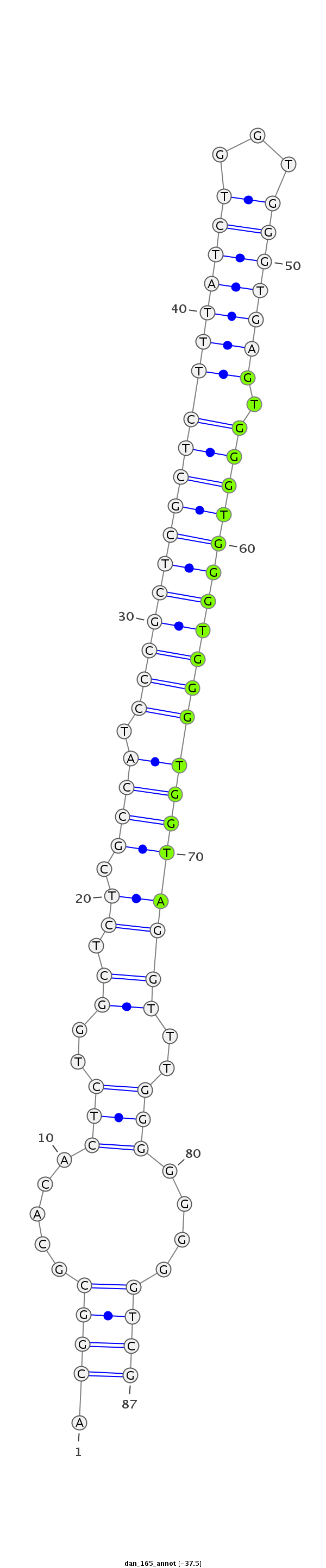
| ........................................................................................GTGGGTGGGTGGGTGGTA................................................... |
18 |
0 |
4 |
42.75 |
171 |
153 |
17 |
0 |
1 |
0 |
| ............................................................................................GTGGGTGGGTGGTAGGGTT.............................................. |
19 |
1 |
4 |
4.50 |
18 |
16 |
0 |
2 |
0 |
0 |
| .......................................................................................AGTGTGTGGGTGGGTGGTAG.................................................. |
20 |
1 |
3 |
1.00 |
3 |
3 |
0 |
0 |
0 |
0 |
| .........................................................................................TGGGAGGGTGGGTAGCAGGTTT.............................................. |
22 |
3 |
1 |
1.00 |
1 |
0 |
0 |
1 |
0 |
0 |
| ............................................................................................GTGGGTGGGTGGTAGGTT............................................... |
18 |
0 |
1 |
1.00 |
1 |
1 |
0 |
0 |
0 |
0 |
| ................................................................................GTGGGTGAGTGGGTGGGTG.......................................................... |
19 |
0 |
8 |
0.75 |
6 |
6 |
0 |
0 |
0 |
0 |
| ................................................................................GTGGGTGAGTGGGTGGGT........................................................... |
18 |
0 |
8 |
0.63 |
5 |
5 |
0 |
0 |
0 |
0 |
| ............................................................................................GTGGGTGGGTGGTAGG................................................. |
16 |
0 |
8 |
0.50 |
4 |
0 |
0 |
4 |
0 |
0 |
| ........................................................................................GTGGGGGGGTGGGTGGTA................................................... |
18 |
1 |
6 |
0.33 |
2 |
2 |
0 |
0 |
0 |
0 |
| ...................................................................................GGTGAGTGGGTGGGTGGGTG...................................................... |
20 |
0 |
4 |
0.25 |
1 |
1 |
0 |
0 |
0 |
0 |
| ........................................................................................GTGGGTGGGTGAGTGGTA................................................... |
18 |
1 |
13 |
0.23 |
3 |
3 |
0 |
0 |
0 |
0 |
| ........................................................................................GTGGGTTGGTGGGTGGTA................................................... |
18 |
1 |
9 |
0.22 |
2 |
2 |
0 |
0 |
0 |
0 |
| ....................................................................................................GTCGTAGGTTAGGGGGGG....................................... |
18 |
2 |
5 |
0.20 |
1 |
0 |
0 |
1 |
0 |
0 |
| ........................................................................................TTGGGTGGGTGGGTGGTA................................................... |
18 |
1 |
12 |
0.17 |
2 |
2 |
0 |
0 |
0 |
0 |
| ............................................................................................GTGGGTGTGTGGTAGGGTT.............................................. |
19 |
2 |
12 |
0.17 |
2 |
2 |
0 |
0 |
0 |
0 |
| .......................................................................................AGTGTGTGGGTTGGTGGTAG.................................................. |
20 |
2 |
7 |
0.14 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...................................................................................................GGTGGGAAGTTTGGCGGGGGT..................................... |
21 |
3 |
7 |
0.14 |
1 |
0 |
1 |
0 |
0 |
0 |
| .................................................................................TGGGTGAGTGGGTGGGTGG......................................................... |
19 |
0 |
7 |
0.14 |
1 |
1 |
0 |
0 |
0 |
0 |
| ............................................................................................GTGGGTGGGTGGTGGGTTT.............................................. |
19 |
1 |
7 |
0.14 |
1 |
0 |
0 |
0 |
1 |
0 |
| ........................................................................................GTGGGTGGGTTGGTGGTA................................................... |
18 |
1 |
8 |
0.13 |
1 |
1 |
0 |
0 |
0 |
0 |
| .................................................................................TGGTTGAGTGGGTGGGCGGGT....................................................... |
21 |
2 |
8 |
0.13 |
1 |
0 |
0 |
1 |
0 |
0 |
| ............................................................................................GTGGGTGGGTCGTAGGGTT.............................................. |
19 |
2 |
8 |
0.13 |
1 |
0 |
1 |
0 |
0 |
0 |
| .......................................................................................................GTAGGTTTGGGGGTGGTA.................................... |
18 |
2 |
9 |
0.11 |
1 |
0 |
0 |
0 |
0 |
1 |
| .......................................................................................................ATAGGGTTGGGGGGGGTC.................................... |
18 |
2 |
9 |
0.11 |
1 |
0 |
0 |
0 |
0 |
1 |
| .............................................................................CTGGTGGGTGAGTAGGCG.............................................................. |
18 |
2 |
10 |
0.10 |
1 |
0 |
0 |
1 |
0 |
0 |
| .........................................................................................TCGGTGGGTGGGTGGTAG.................................................. |
18 |
1 |
10 |
0.10 |
1 |
1 |
0 |
0 |
0 |
0 |
| .........................................................................................TGGGAGTGTGGCTGGTAGGTT............................................... |
21 |
3 |
10 |
0.10 |
1 |
0 |
0 |
1 |
0 |
0 |
| ........................................................................................GTGGGTGGGTGGGAGGTA................................................... |
18 |
1 |
10 |
0.10 |
1 |
0 |
1 |
0 |
0 |
0 |
| .................................................................................................TGGGTGGAAGGTTTGGGC.......................................... |
18 |
2 |
10 |
0.10 |
1 |
0 |
0 |
1 |
0 |
0 |
| ....................................................................................GTGAGTGGGTGGGTGGGT....................................................... |
18 |
0 |
12 |
0.08 |
1 |
1 |
0 |
0 |
0 |
0 |
| .................................................................................TGGCTGAGTGCGTGGGTGGGT....................................................... |
21 |
2 |
12 |
0.08 |
1 |
0 |
0 |
1 |
0 |
0 |
| ........................................................................................GTGGTTGGGTGGGTGGTA................................................... |
18 |
1 |
14 |
0.07 |
1 |
1 |
0 |
0 |
0 |
0 |
| ................................................................................GTTGGTGAGTGGGTGGGT........................................................... |
18 |
1 |
15 |
0.07 |
1 |
1 |
0 |
0 |
0 |
0 |
| ............................................................................................................ATTGGGGGGGGTGGCTGGCC............................. |
20 |
3 |
20 |
0.05 |
1 |
0 |
0 |
1 |
0 |
0 |
| ........................................................................................GTGGGTGGGTGGGGGGTAA.................................................. |
19 |
2 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
| ................................................................................GTGGGTGATTGGGTGGGT........................................................... |
18 |
1 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
| ...................................................................................GCTGAGAGGGTGGGTGCGTGG..................................................... |
21 |
3 |
20 |
0.05 |
1 |
0 |
0 |
0 |
0 |
1 |
| .........................................................................................TCGTTGGGTGGGTGGTAG.................................................. |
18 |
2 |
20 |
0.05 |
1 |
1 |
0 |
0 |
0 |
0 |
| .................................................................................TGGGTGAGTGGCTAGCTGGGT....................................................... |
21 |
3 |
20 |
0.05 |
1 |
0 |
0 |
1 |
0 |
0 |