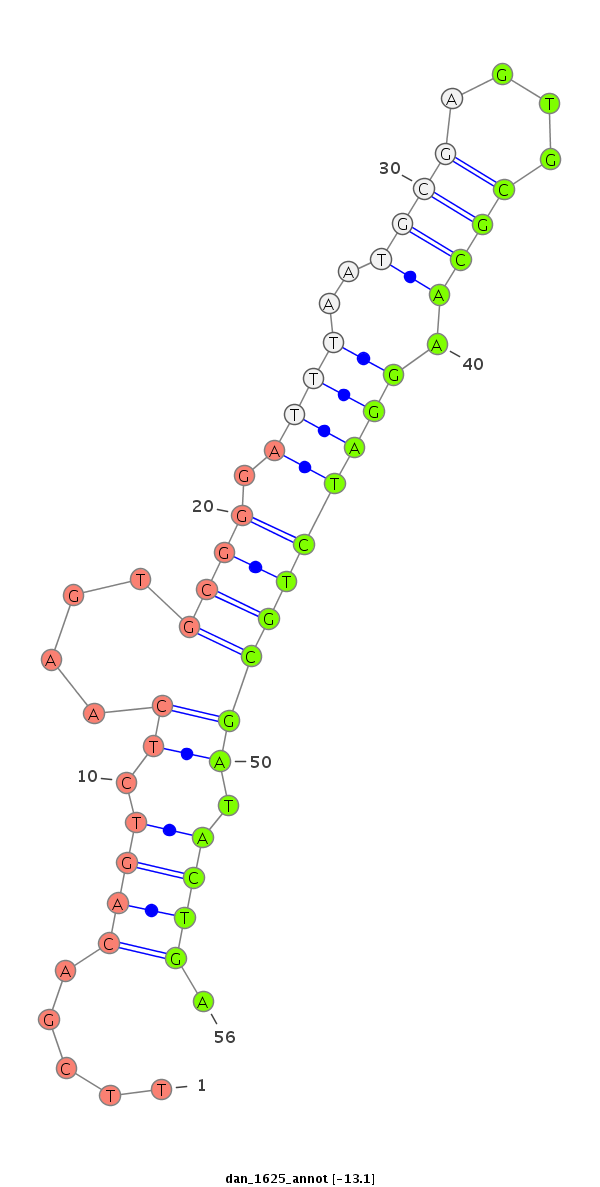



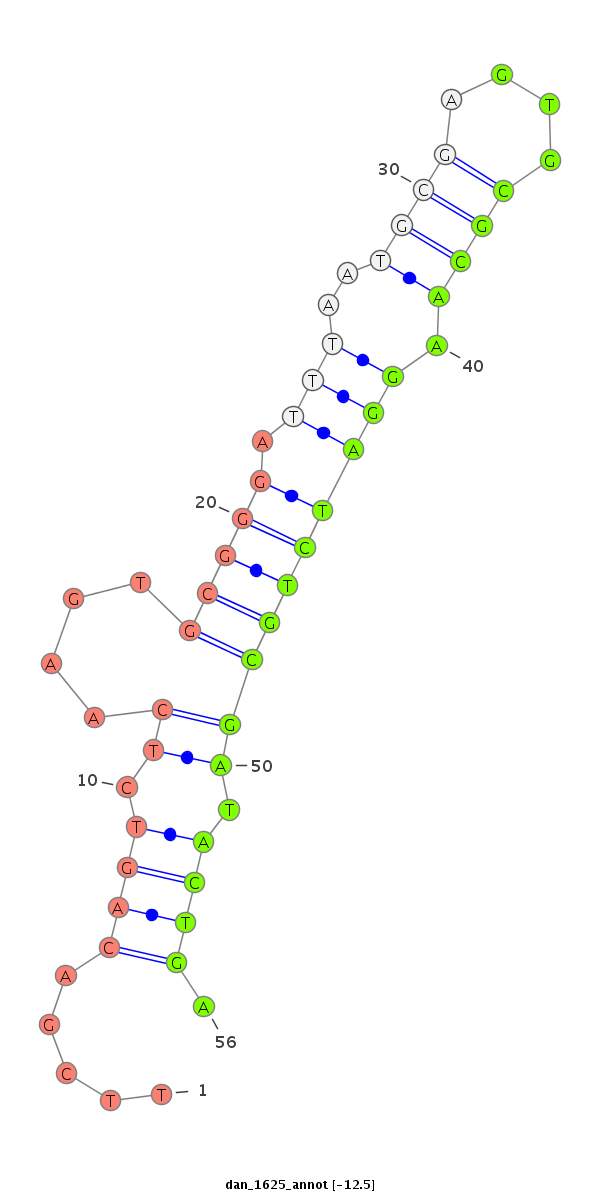
ID:dan_1625 |
Coordinate:scaffold_13250:3358454-3358604 - |
Confidence:Candidate |
Type:Unknown |
[View on UCSC Genome Browser {Cornell Mirror}] |
| Legend: | mature | star | mismatch in alignment | mismatch in read |
 |
 |
 |
 |
 |
| -13.1 | -13.1 | -12.5 |
 |
 |
 |
exon [dana_GLEANR_1009:2]; CDS [Dana\GF10133-cds]; intron [Dana\GF10133-in]; Antisense to intron [Dana\GF12903-in]
No Repeatable elements found
| mature | star |
| ##################################################--------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- GAAGGAACTCCCACCTTCTAGTGGGGAATAGGCTGAAGGACCCCCCGTGGGTAAGTTCGACAGTCTCAAGTGCGGGATTTAATGCGAGTGCGCAAGGATCTGCGATACTGATCGCAGCTACTGAGCTTAAGGACCCCCTGTGGGTAAGTCCGGCGGCGGTACGCGAATCAGCAAAAGAGTATAAATCAAGGAGCAAGTGGGAAAAGGATCAAGGATCCGCGGTGGAAATGAAGCCCATGGGTAAAACGAGT *******************************************************.....((((.((....((((.((((..((((....)))).)))))))))).)))).******************************************************************************************************************************************** |
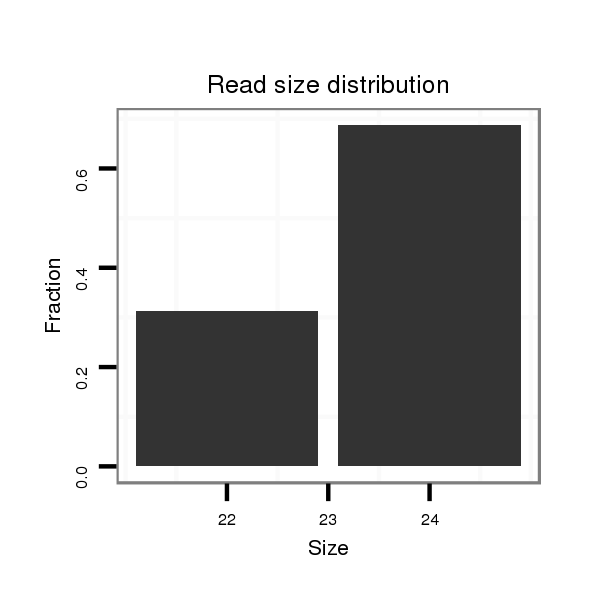
Read size | # Mismatch | Hit Count | Total Norm | Total | M044 female body |
M058 embryo |
V106 head |
V039 embryo |
V105 male body |
V055 head |
|---|---|---|---|---|---|---|---|---|---|---|---|
| ....................................................................................................................................................................................................................GGTTCCGGGGTCGAAATGA.................... | 19 | 3 | 18 | 2.11 | 38 | 29 | 0 | 6 | 0 | 2 | 1 |
| ...................................................................................................................................................................................................................AGGTTCCGGGGTCGAAATGA.................... | 20 | 3 | 4 | 1.75 | 7 | 6 | 0 | 1 | 0 | 0 | 0 |
| ...................................................................................................................................................................................................................AGGTTCCGGGGTCGAAATG..................... | 19 | 3 | 12 | 1.00 | 12 | 8 | 0 | 0 | 0 | 3 | 1 |
| ...................................................................................................................................................................................................................AGGTTCCGCGGTCGAAATGAC................... | 21 | 3 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................AAGGATCCGCGGTGGAAATGAAGCCC............... | 26 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................GTGCGCAAGGATCTGCGATACTGA............................................................................................................................................ | 24 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .............................................................................................AAGGATCTGCGATACTTATCGCAGC..................................................................................................................................... | 25 | 1 | 2 | 0.50 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| .....................................................................................................................................................................................TAAATCAAGGAGCAAGTGGGA................................................. | 21 | 0 | 4 | 0.25 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .......................................................................................................................ACTGAGATGTAGGACCCCCTGT.............................................................................................................. | 22 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| ....................GTGGGGTATGGGCTGAAG..................................................................................................................................................................................................................... | 18 | 2 | 10 | 0.20 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| .......................................................TTCGACAGTCTCAAGTGCGGGA.............................................................................................................................................................................. | 22 | 0 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................AAAGGATCAAGGATCCGCGGTG........................... | 22 | 0 | 13 | 0.15 | 2 | 2 | 0 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................................................................................GAGGTTCCGCGGTCGAAAT...................... | 19 | 3 | 9 | 0.11 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| .........................................................................................................................................................................................................AAAAGGATCAAGGATCCGCGGTGG.......................... | 24 | 0 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................................AAAGGATCAAGGATCCGCGGTGGAA........................ | 25 | 0 | 13 | 0.08 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................................AGGATCAAGGATCCGCGGTGGA......................... | 22 | 0 | 15 | 0.07 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .........................................................................................................................................................................................................AAAAGGATCAAGGATCCGCGGT............................ | 22 | 0 | 15 | 0.07 | 1 | 0 | 0 | 0 | 0 | 1 | 0 |
| ......................................................................................................................................................................................................................ATCAGGGGTGGAAATGA.................... | 17 | 2 | 20 | 0.05 | 1 | 0 | 0 | 0 | 0 | 0 | 1 |
| .................................................................................................................................................................................................CAAGTGGGAAAAGGATCAAGGATCCGC............................... | 27 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ............................TAGGCTGAAGGACCCCCCGTGGGTAAG.................................................................................................................................................................................................... | 27 | 0 | 20 | 0.05 | 1 | 0 | 0 | 0 | 1 | 0 | 0 |
| ...................................................................................................................................................................................................AGTGGGAAAAGGATCAAGGATCC................................. | 23 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................AAGTCCGGCGGCGGTACGCGAA.................................................................................... | 22 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ................................................................................................................................................TAAGTCCGTCGGCGGTACGCGAATCAGC............................................................................... | 28 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................TTAAGGACCCCCTGTGGGTAAGTC..................................................................................................... | 24 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 | 0 |
|
CTTCCTTGAGGGTGGAAGATCACCCCTTATCCGACTTCCTGGGGGGCACCCATTCAAGCTGTCAGAGTTCACGCCCTAAATTACGCTCACGCGTTCCTAGACGCTATGACTAGCGTCGATGACTCGAATTCCTGGGGGACACCCATTCAGGCCGCCGCCATGCGCTTAGTCGTTTTCTCATATTTAGTTCCTCGTTCACCCTTTTCCTAGTTCCTAGGCGCCACCTTTACTTCGGGTACCCATTTTGCTCA
********************************************************************************************************************************************.....((((.((....((((.((((..((((....)))).)))))))))).)))).******************************************************* |
Read size | # Mismatch | Hit Count | Total Norm | Total | M044 female body |
M058 embryo |
V039 embryo |
V055 head |
V106 head |
|---|---|---|---|---|---|---|---|---|---|---|
| ............TGGAAGATCACCCCTTATCCGACTTC..................................................................................................................................................................................................................... | 26 | 0 | 1 | 1.00 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..........................TTATCCGACTTCCTGGGGGGCACCCAT...................................................................................................................................................................................................... | 27 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................CGCCGCCATGCGCTTAGTCGTT............................................................................. | 22 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................CAGGCCGCCGCCATGCGCTTAGTCGTTT............................................................................ | 28 | 0 | 1 | 1.00 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................CCATTCAGGCCGCCGCCATGCGCTT.................................................................................... | 25 | 0 | 20 | 0.70 | 14 | 0 | 14 | 0 | 0 | 0 |
| ................................................................................TTACGCTCACGCGTTCCTAGACGCTAT................................................................................................................................................ | 27 | 0 | 2 | 0.50 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................CCGCCATGCGCTTAGTCGTT............................................................................. | 20 | 0 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| ......................................................................................................................................................GCCGCCGCCATGAGCTTAGTCGTT............................................................................. | 24 | 1 | 2 | 0.50 | 1 | 0 | 1 | 0 | 0 | 0 |
| .....................................................TCAAGCTGTCAGAGTTCACGCCCTAAAT.......................................................................................................................................................................... | 28 | 0 | 5 | 0.40 | 2 | 2 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................GTTCCTCGTTCACCCTTTTCCTAGTT....................................... | 26 | 0 | 20 | 0.35 | 7 | 7 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................CCTCGTTCACCCTTTTCCTAGTTCCT.................................... | 26 | 0 | 20 | 0.30 | 6 | 0 | 6 | 0 | 0 | 0 |
| .............................................................................................................................................CCCATTCAGGCCGCCGCCATGCGC...................................................................................... | 24 | 0 | 20 | 0.30 | 6 | 6 | 0 | 0 | 0 | 0 |
| ..................................................................................................AGACGCTATGACTCCCGTTG..................................................................................................................................... | 20 | 3 | 4 | 0.25 | 1 | 0 | 0 | 0 | 1 | 0 |
| ..........................................................................................................................CTCGAATTCCTGGGGGACATCC........................................................................................................... | 22 | 1 | 4 | 0.25 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................CGGGGGGCACCCATTCAAGCTGTCAGAG........................................................................................................................................................................................ | 28 | 1 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................................................................................................................................................................AGTTCCTCGTTCACCCTTTTCCTAGTT....................................... | 27 | 0 | 20 | 0.20 | 4 | 3 | 1 | 0 | 0 | 0 |
| ..................................................................................................................................................TCAGGCCGCCGCCATGCGCTTAGTC................................................................................ | 25 | 0 | 20 | 0.20 | 4 | 4 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................GGGGGACACCCATTCAGGCCGCCGCCA........................................................................................... | 27 | 0 | 20 | 0.20 | 4 | 4 | 0 | 0 | 0 | 0 |
| .....................................................................CACGCCCTAAATTACGCTCACGCGT............................................................................................................................................................. | 25 | 0 | 5 | 0.20 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................GGGCACCCATTCAAGCTGTCAG.......................................................................................................................................................................................... | 22 | 0 | 11 | 0.18 | 2 | 2 | 0 | 0 | 0 | 0 |
| ...........................................GGGCACCCATTCAAGCTGTCATAGT....................................................................................................................................................................................... | 25 | 1 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 |
| ............................................................GTCAGAGTTCACGCCCTAAATTACGCT.................................................................................................................................................................... | 27 | 0 | 6 | 0.17 | 1 | 1 | 0 | 0 | 0 | 0 |
| ....................................................................TCACGCCCTAAATTACGCTCACG................................................................................................................................................................ | 23 | 0 | 6 | 0.17 | 1 | 0 | 1 | 0 | 0 | 0 |
| .............................................................................................................................................CCCATTCAGGCCGCCGCCATGCG....................................................................................... | 23 | 0 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................................................................TAGTTCCTCGTTCACCCTTTTCCTAGTT....................................... | 28 | 0 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 | 0 |
| .............................................................................................................................................................................................CCTCGTTCACCCTTTTCCTAGTTC...................................... | 24 | 0 | 20 | 0.15 | 3 | 1 | 2 | 0 | 0 | 0 |
| .............................................................................................................................................CCCATTCAGGCCGCCGCCATG......................................................................................... | 21 | 0 | 20 | 0.15 | 3 | 3 | 0 | 0 | 0 | 0 |
| ...................................................................................................................................................CAGGCCGCCGCCATGCGCTTAGT................................................................................. | 23 | 0 | 20 | 0.15 | 3 | 2 | 0 | 1 | 0 | 0 |
| ...................................................................................................................................................CAGGCCGCCGCCATGCGCTTAGTC................................................................................ | 24 | 0 | 20 | 0.15 | 3 | 2 | 1 | 0 | 0 | 0 |
| ...............................................ACCCATTCAAGCTGTCAGAGTTCAC................................................................................................................................................................................... | 25 | 0 | 7 | 0.14 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................GCCACCCATTCAAGCTGTCAGAGT....................................................................................................................................................................................... | 24 | 1 | 7 | 0.14 | 1 | 0 | 0 | 1 | 0 | 0 |
| ..........................................................................................................................CTCGAAATCCTGTGGGAC............................................................................................................... | 18 | 2 | 7 | 0.14 | 1 | 0 | 0 | 0 | 0 | 1 |
| .......................................................AAGCTGTCAGAGTTCACGC................................................................................................................................................................................. | 19 | 0 | 8 | 0.13 | 1 | 0 | 1 | 0 | 0 | 0 |
| .......................................TGGGGGGCACCCATTCAAGCTGTCAG.......................................................................................................................................................................................... | 26 | 0 | 8 | 0.13 | 1 | 1 | 0 | 0 | 0 | 0 |
| .........................................GGGGGCACCCATTCAAGCTGTCAG.......................................................................................................................................................................................... | 24 | 0 | 9 | 0.11 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................ACACCCATTCAGGCCGCCGCCATGC........................................................................................ | 25 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................TCCTCGTTCACCCTTTTCCTAGTTC...................................... | 25 | 0 | 20 | 0.10 | 2 | 1 | 0 | 1 | 0 | 0 |
| .........................................GGGGGCACCCATTCAAGCTGT............................................................................................................................................................................................. | 21 | 0 | 10 | 0.10 | 1 | 1 | 0 | 0 | 0 | 0 |
| ........................................................................................................................................GGACACCCATTCAGGCCGCCGCCATGCGC...................................................................................... | 29 | 0 | 20 | 0.10 | 2 | 2 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................................CCTTTTCCTAGTTCCTAGGCGCCAC........................... | 25 | 0 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 |
| ......................................................................................................................................................................................................CCCTTTTCCTAGTTCCTAGGCGCCAC........................... | 26 | 0 | 13 | 0.08 | 1 | 1 | 0 | 0 | 0 | 0 |
| .......................................................................................................................................................................................TTAGTTCCTCGTTCACCCTTTTCCTAGT........................................ | 28 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................ACACCCATTCAGGCCGCCGCCA........................................................................................... | 22 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................TTCCTCGTTCACCCTTTTCCTAGTT....................................... | 25 | 0 | 20 | 0.05 | 1 | 0 | 0 | 1 | 0 | 0 |
| ......................................................................................................................................GGGGACACCCATTCAGGCCGCCTC............................................................................................. | 24 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..........................................................CTGTCAGAGTTCACGCCCTAAAT.......................................................................................................................................................................... | 23 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................CACCCATTCAGGCCGCCGCCATGCGCTT.................................................................................... | 28 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................TTCCTCGTTCACCCTTTTCCTAGT........................................ | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................TTCCTCGTTCACCCTTATCCTAGTT....................................... | 25 | 1 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..............................................................................................................................................................................................CTCGTTCACCCTTTTCCTAGTTCATA................................... | 26 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .........................................................GCTGTCAGAGTTCACGCCCT.............................................................................................................................................................................. | 20 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................................................................TCCTCGTTCACCCTTTTCCTAGTT....................................... | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .................................................................................................................................................TTCAGGCCGCCGCCATGCGCTTAGTC................................................................................ | 26 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..................................................................................................................................................CCAGGCCGCCGCCATGCGCTTAGT................................................................................. | 24 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| .................................................................................................................................................TTCAGGCCGCCGCCATGCGCTTAGGC................................................................................ | 26 | 1 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ..............................................................................................................................................CCATTCAGGCCGCCGCCATGCGC...................................................................................... | 23 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ..........................................................................................................................................................................................GTTCCTCGTTCACCCTTTTCCTAGTTCC..................................... | 28 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ............................................................................................................................................ACCCATTCAGGCCGCCGCCATGCGCTTAG.................................................................................. | 29 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| .....................................................................................................................................GGGGGACACCCATTCAGGCCGCCGCCATG......................................................................................... | 29 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ...........................................................................................................................................................................................TTCCTCGTTCACCCTTTTCCT........................................... | 21 | 0 | 20 | 0.05 | 1 | 0 | 1 | 0 | 0 | 0 |
| ................................................................................................................................................ATTCAGGCCGCCGCCATGCGCTTA................................................................................... | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| ...........................................................................................................................................CACCCATTCAGGCCGCCGCCATGC........................................................................................ | 24 | 0 | 20 | 0.05 | 1 | 1 | 0 | 0 | 0 | 0 |
| Species | Coordinate | ID | Alignment |
|---|---|---|---|
| droAna3 | scaffold_13250:3358404-3358654 - | dan_1625 | GAAGGAACTCCCACCTTCTAGTGGGGAATAGGCTGAAGGACCCCCCGTGGGTAAGTTCGACAGTC------TCAAGTGCGGGATTTAATGCGAGTGCGCAAGGATCTGCGATACTGATCGCAGCTAC-TGAGCTTAAGGACCCCCTGTGGGTAAGTCCGGCGGCGGTACGCGAATCAGCAAAAG---AGTATAAATCAAGGAGCAAGTGGGAAAAGGATCAAGGATCCGCGGTGGAAATG------------AAGCCCATGGGTAAAACGAGT |
| droBip1 | scf7180000396486:4218-4430 + | CTTGTAGCGTT--------------GAGTAGG----AGGACCGCC-TTTGGAAAGTCCGACATTCAAGTGTTCAAGAGCGACATTCAA-----GAGCG-------TTTCTAAAAAGGACGCGGTTATTTGAGTTGAAGGACCGCCTTTGA-AGAGTCCAACA---------------GCCCAAAGAAAGGAGAACGTAAAGAGCGAGTGA---AAG-------GATCCGTTGCAGGA-TGGTTCCCCAGGACAAGCCCAGG-GTATCA-GAGT | |
| droFic1 | scf7180000453342:66121-66160 - | CA-------------------------------------------------------------------------------------------------------------------------------------------------------------------------------GCGAAAA---AAAATAAGTAGAGGAGCAAGGGAGGAGAGGA------------------------------------------------------- | |
| droEug1 | scf7180000409905:57628-57642 + | GAGGCAACTCCCACC------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------ | |
| droGri2 | scaffold_15110:21448829-21448841 + | GGT---ACTCATACCT----------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------------- |
| Species | Read alignment | |||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| droAna3 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droBip1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droFic1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droEug1 |
|
|||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
| droGri2 |
|
Generated: 05/18/2015 at 02:33 AM